Dr Rajiv Desai
An Educational Blog
ANTIMICROBIAL RESISTANCE (AMR)
Antimicrobial resistance (AMR):
______
______
Prologue:
Within a few days of scraping his leg in a scooter accident in 2009, nine-year-old Brock Wade was in hospital fighting for his life with a methicillin-resistant staphylococcus aureus (MRSA) infection. Once the infection – caused by one of the bacteria most often resistant to antibiotics – has been diagnosed, doctors put him on five different antibiotics. After a month in the hospital, and against all odds, Brock recovered and was well enough to come home. Scenarios such as this case are increasingly being played out all over the world. But not all the thousands of patients that contract drug-resistant bacterial infections every year are as lucky as Brock. And the problem looks set to get worse. While infectious agents are becoming more and more resistant to the medicines that are currently in use, not enough drugs are being developed to combat them. WHO defines antimicrobial resistance (AMR) as a microorganism’s resistance to an antimicrobial drug that was once able to treat an infection by that microorganism. AMR threatens to make existing standard medical treatments ineffective. This means that currently common and treatable infections are becoming life threatening. WHO describes AMR as one of the greatest threats currently facing human health. AMR is present in all parts of the world affecting humans, animals and agriculture. Without urgent coordinated action, the world is heading towards a post-antibiotic era, in which common infections and minor injuries, which have been treatable for decades, can once again kill. The whole of modern healthcare including invasive surgery and immunosuppressive chemotherapy is facing threat of AMR. Homo-sapiens is an alien species on earth. This planet belongs to bacteria. There are more bacteria on earth than all other living organisms. The human body contains more number of bacteria than human cells themselves. We lived with arrogant optimism that we had conquered infections, at least the bacterial infections, if not the viruses. How wrong we were! Bacteria have finally reclaimed their premier status and superiority and won the war against humans. They are literally mocking our intellect, knowledge and antibiotic weaponry. The introduction of penicillin dramatically changed the health outcomes of patients with bacteria-induced pneumonia and bloodstream infection from a case fatality rate of about 90% to a survival rate of about 90%. Sir Alexander Fleming discovered the first antibiotic penicillin in 1928. In his Nobel Prize lecture on December 11, 1945, Fleming said ‘It is not difficult to make microbes resistant to penicillin in the laboratory by exposing them to concentrations not sufficient to kill them…there is the danger that the ignorant man may easily under-dose himself and by exposing his microbes to non-lethal quantities of the drug make them resistant.’ He continued, ‘Mr X has a sore throat. He buys some penicillin and gives himself, not enough to kill the streptococci but enough to educate them to resist penicillin. He then infects his wife. Mrs X gets pneumonia and is treated with penicillin. As the streptococci are now resistant to penicillin the treatment fails. Mrs X dies.’ This example was used to warn colleagues and future generations on the unavoidable consequences derived by an inappropriate use of the antibiotic he had discovered. Better than anything else, these words provide an accurate description of the threat imposed by AMR.
____
Note:
This article is written to create awareness about AMR and to find ways to reduce/overcome AMR. Disease caused by every resistant microorganism and its treatment is beyond the scope of this article.
____
Abbreviations and synonyms:
AMTs = Antimicrobial therapies
AMR = Antimicrobial resistance
ARMs = Antimicrobial-resistant microorganisms
MDR = multidrug resistance
GNB = gram negative bacteria
PBP = penicillin binding protein
r gene = resistance gene
HGT = horizontal gene transfer
ESBL= extended-spectrum beta-lactamases
ASP = antimicrobial stewardship programs
____
____
Nomenclature of antibiotics, antimicrobials and antimicrobial resistance (AMR):
_
Microorganisms:
Microorganisms are very tiny living things. They are so small that you need a microscope to see them. A microorganism or microbe is a microscopic living organism, which may be single-celled or multicellular. Microorganisms are very diverse and include all bacteria, archaea, most protozoa and some species of fungi & algae. Viruses are generally regarded as not living but some microbiologists also classify viruses as microorganisms. You might see with your naked eye a group of these organisms – such as a mold growth on bread – but you can’t see each individual fungal cell without a microscope.
_
Antibiotic:
The term antibiotic was first used in 1942 by Selman Waksman and his collaborators in journal articles to describe any substance produced by a microorganism that is antagonistic to the growth of other microorganisms in high dilution. This definition excluded substances that kill bacteria but that are not produced by microorganisms (such as gastric juices and hydrogen peroxide). It also excluded synthetic antibacterial compounds such as the sulfonamides. In current usage, the term “antibiotic” is applied to any medication that kills bacteria or inhibits their growth, regardless of whether that medication is produced by a microorganism or not. Therefore generic term “antibiotic” is used here to denote any class of organic molecule that inhibits or kills microbes by specific interactions with bacterial targets, without any consideration of the source of the particular compound or class. Thus, purely synthetic therapeutics are considered antibiotics; after all, they interact with receptors and provoke specific cell responses and biochemical mechanisms of cross-resistance in pathogens. The fluoroquinolones, sulfonamides, and trimethoprim are good examples. Antibiotics can be further subdivided into bactericidal agents, which kill bacteria, and bacteriostatic agents, which slow down or stall bacterial growth. Antibiotics are also subdivided into two categories, broad spectrum and narrow spectrum, based on the number and types of bacteria they affect. Broad spectrum antibiotics are effective against many types of bacteria, while narrow spectrum antibiotics are effective against a more limited range of bacteria.
_
Antimicrobial:
An antimicrobial is an agent that kills microorganisms or inhibits their growth. Antimicrobials are active substances of synthetic or natural origin which destroy microorganisms (bacteria, viruses, fungi and parasites), suppressing their growth or their ability to reproduce in animals or humans. Antimicrobial medicines can be grouped according to the microorganisms they act primarily against. For example, antibiotics are used against bacteria and antifungals are used against fungi. All antibiotics are antimicrobials, but not all antimicrobials are antibiotics. For example, anti-viral drugs and anti-fungal drugs are antimicrobials, but they are not antibiotics. They can also be classified according to their function. Agents that kill microbes are called microbicidal, while those that merely inhibit their growth are called biostatic. The use of antimicrobial medicines to treat infection is known as antimicrobial chemotherapy, while the use of antimicrobial medicines to prevent infection is known as antimicrobial prophylaxis.
_
The main classes of antimicrobial agents are disinfectants (“nonselective antimicrobials” such as bleach), which kill a wide range of microbes on non-living surfaces to prevent the spread of illness, antiseptics (which are applied to living tissue and help reduce infection during surgery), and antibiotics (which destroy microorganisms within the body). Antiseptics are antimicrobial substances that are applied to living tissue/skin to reduce the possibility of infection, sepsis, or putrefaction. Antiseptics are generally distinguished from antibiotics by the latter’s ability to be transported through the circulatory system to destroy bacteria within the body, and from disinfectants, which destroy microorganisms found on non-living objects. Antiseptics are typically small simple molecules such as ethanol, or soap, or phenol, that kill all types of cells (including your own) by non-specific mechanisms, such as disrupting cell membranes. They are never taken internally, as they are poisons. Antibiotics, whether applied systemically or topically, are more-complex molecules that bind to specific molecular targets in bacteria and disrupt their functioning. Because they bind to microbial proteins, and not human ones, they have very low toxicity. Disinfectants do not kill bacterial spores e.g., on surgical instruments; a sterilization process is required for that. Even sterilization may not destroy prions. The researchers found that hypochlorous acid, the active ingredient in bleach, causes the unfolding of proteins in bacteria in much the same was that heat stress or fever does. Those denatured proteins then clump together irreversibly into a mass in living cells, similar to what happens to proteins when you boil an egg.
_
Antimicrobial resistance (AMR):
Antibiotics are effective against bacteria, just one class of micro-organism, while the term antimicrobial resistance (AMR) covers the development of resistance in all micro-organisms—including bacteria, fungi, viruses and protozoa (such as the parasite that causes malaria)—to the various agents used to treat the infections they cause. Antimicrobial resistance is the ability of a microorganism to stop an antimicrobial (such as antibiotics, antivirals and antimalarials) from working against it. As a result, standard treatments become ineffective, infections persist and may spread to others. The WHO defines antimicrobial resistance as a microorganism’s resistance to an antimicrobial drug that was once able to treat an infection by that microorganism. A person cannot become resistant to antibiotics. Resistance is a property of the microbe, not a person or other organism infected by a microbe.
The figure above shows difference between non-resistant bacteria and drug resistant bacteria. Non-resistant bacteria multiply, and upon drug treatment, the bacteria die. Drug resistant bacteria multiply as well, but upon drug treatment, the bacteria continue to spread. AMR is the ability of a microorganism strain to survive and/or to multiply despite the administration and absorption of a drug given in doses equal or higher than those usually recommended, but within the limits of tolerance of the patients. The term is antimicrobial resistance refers to strains of bacteria and other pathogens that have mutated so that they are resistant to antibiotics. Antimicrobial resistance is when bacteria or other microbes become resistant to the effects of a drug after being exposed to it. This means that the drug, and similar drugs, will no longer be effective against those microbes. Antimicrobial resistance is a complex phenomenon with many causes. We know that all uses of antimicrobials, whether in humans or animals, can spur resistance. Sometimes resistance even occurs spontaneously. The worst-case AMR nightmare is a plague of superbugs that are resistant to every known antibiotic. There aren’t any of those yet but many strains have been found that are resistant to most of the antibiotics that are commonly used. Bacteria that are resistant to a large number of antibiotics or are resistant to the antibiotics that are used when all else fails are called superbugs.
_
Susceptibility and resistance:
The term “susceptible” simply means that the microorganism is capable of being affected by the antimicrobial. For example, if we say that a type of Streptococcus bacteria is susceptible to penicillin, it means that the bacteria are killed or growth is inhibited (stopped) by the penicillin. Antimicrobial resistance (as well as antibiotic resistance) occurs when a microorganism develops the ability to resist the action of an antimicrobial. Basically, the microorganism develops the ability to survive and reproduce in the presence (and dose) of an antimicrobial that used to prevent these actions. In general, it’s really only considered “resistance” when it occurs in an organism that used to be susceptible to an antimicrobial’s effects but now is not susceptible; this doesn’t really apply to an organism that was never susceptible to that antimicrobial. How resistance develops is a very complex process, and we don’t really know all of the factors or events that can make it happen. We do know that an organism can undergo a change in its DNA that makes it resistant to one or more antimicrobials – this change might just be an accident that turns out to be fortunate for that organism, or it might be in response to something else, such as the use of antimicrobials – and it might transfer that changed DNA to another organism or pass it on to its offspring when it reproduces. The term ‘antibiotic resistome’ was proposed for the collection of all antibiotic resistance genes in microorganisms, from both pathogenic and nonpathogenic bacteria. The term ‘microbiome’ has been suggested by Nobel laureate Joshua Lederberg to describe the collective genome of our indigenous microbes.
_________
_________
Introduction to antimicrobial resistance (AMR):
_
We use antibiotics throughout our lifetime:
_
Avoiding infection has always been expensive. Some human populations escaped tropical infections by migrating into cold climates but then had to procure fuel, warm clothing, durable housing, and crops from a short growing season. Waterborne infections were averted by owning your own well or supporting a community reservoir. Everyone got vaccines in rich countries, while people in others got them later if at all. Antimicrobial agents seemed at first to be an exception. They did not need to be delivered through a cold chain and to everyone, as vaccines did. They had to be given only to infected patients and often then as relatively cheap injectables or pills off a shelf for only a few days to get astonishing cures. Antimicrobials not only were better than most other innovations but also reached more of the world’s people sooner. The problem appeared later. After each new antimicrobial became widely used, genes expressing resistance to it began to emerge and spread through bacterial populations. Patients infected with bacteria expressing such resistance genes then failed treatment and remained infected or died. Growing resistance to antimicrobial agents began to take away more and more of the cures that the agents had brought.
_
Benefits of Antibiotics:
For an American in the 21st century, it is hard to imagine the world before antibiotics. At the beginning of the 20th century, as many as nine women out of every 1,000 who gave birth died, 40 percent from sepsis. In some cities as many as 30 percent of children died before their first birthday. One of every nine people who developed a serious skin infection died, even from something as simple as a scrape or an insect bite. Pneumonia killed 30 percent of those who contracted it; meningitis killed 70 percent. Ear infections caused deafness; sore throats were not infrequently followed by rheumatic fever and heart failure. Surgical procedures were associated with high morbidity and mortality due to infection. Of course, the statistics from third world is worse than American statistics. This picture changed dramatically with three major developments: improvements in public health, vaccines, and antibiotics. Over the course of the 20th century, deaths from infectious diseases declined markedly and contributed to a substantial increase in life expectancy. Antibiotics, in particular, have saved millions of lives. Antibiotics have not only saved patients’ lives, they have played a pivotal role in achieving major advances in medicine and surgery. They have successfully prevented or treated infections that can occur in patients who are receiving chemotherapy treatments; who have chronic diseases such as diabetes, end-stage renal disease, or rheumatoid arthritis; or who have had complex surgeries such as organ transplants, joint replacements, or cardiac surgery. Antibiotics have also helped to extend expected life spans by changing the outcome of bacterial infections. In 1920, people in the U.S. were expected to live to be only 56.4 years old; now, however, the average U.S. life span is nearly 80 years. Antibiotics have had similar beneficial effects worldwide. In developing countries where sanitation is still poor, antibiotics decrease the morbidity and mortality caused by food-borne and other poverty-related infections. The World Health Organization ranking of antimicrobials according to their relative importance in human medicine was recently updated. Antimicrobials considered the highest priority among the critically important antimicrobials were quinolones, third- and fourth-generation cephalosporins, macrolides and ketolides, and glycopeptides.
_
Infections caused by microorganisms have threatened human life since time immemorial. During the pre-antibiotic era, these have been a major concern for the high morbidity and mortality in humans. Some of the virulent organisms with the potential to spread infection from one infected person to another at a very rapid rate may cause worldwide pandemics, epidemics or outbreaks. With the discovery of the first antibiotic, “the magic bullet” Penicillin in the year 1943, patients could be effectively cured of many life-threatening infections. This gave a huge relief to the medical practitioners. Next three decades saw the development and discovery of a wide variety of antimicrobial agents. Subsequently, the pace of discovery of newer molecules declined from 1970 to 1987. It has reached a “discovery void” level from 1987 onwards up till now. Antimicrobial agents represent one of the main therapeutic tools both in human and veterinary medicine to control and treat a variety of bacterial infectious diseases. However, during the past five decades, the use and sometimes misuse of antimicrobials in both human and veterinary medicine has resulted in the emergence of strains of bacteria that no longer respond to antimicrobial therapy. Not only do antimicrobial-resistant bacterial pathogens in animals pose a risk in terms of animal health, they also affect public health when transmitted to humans as foodborne contaminants. Thus, addressing the issue of antimicrobial resistance is one of the most urgent priorities in the fields of public health today.
_
Microorganisms have existed on the earth for more than 3.8 billion years and exhibit the greatest genetic and metabolic diversity. They are an essential component of the biosphere and serve an important role in the maintenance and sustainability of ecosystems. Micro-organisms are a global public good. They made Earth liveable before humans evolved and they continue to do so. They do this not only by being the engines that run the critical nutrient cycles, secure air, water, and soil qualities, but also by helping our bodies’ digestive and immune systems develop. It is believed that they compose about 60% of the living biomass. In order to survive, they have evolved mechanisms that enable them to respond to selective pressure exerted by various environments and competitive challenges. The disease-causing microorganisms have particularly been vulnerable to man’s selfishness for survival who has sought to deprive them of their habitat using antimicrobial agents. These microorganisms have responded by developing resistance mechanisms to fight off this offensive. Currently antimicrobial resistance among bacteria, viruses, parasites, and other disease-causing organisms is a serious threat to infectious disease management globally. There is nothing new under the sun, least of all antimicrobial resistance (AMR). Microbes that are antibiotic producers have always needed to be resistant to their own antibiotic. What is rapidly changing, however, is the scale of this resistance and its impact on human beings. Microbes have globalized along with their hosts, while at the same time antimicrobial consumption by these hosts—both humans and animals—has exploded. The gene pool for antimicrobial resistance has never been so accessible, nor its selection pressure so strong. Resistance arises through one of three ways: natural resistance in certain types of bacteria; genetic mutation; or by one species acquiring resistance from another. Resistance can appear spontaneously because of random mutations; or more commonly following gradual buildup over time, and because of misuse of antibiotics or antimicrobials.
_
There has probably been a gene pool in nature for resistance to antibiotic as long as there has been for antibiotic production, for most microbes that are antibiotic producers are resistant to their own antibiotic. In retrospect, it is not surprising that resistance to penicillin in some strains of staphylococci was recognized almost immediately after introduction of the drug. Likewise, very soon after their introduction in the late 1940s, resistance to streptomycin, chloramphenicol and tetracycline was noted. By 1953, during a Shigella outbreak in Japan, a strain of the dysentery bacillus (Shigella dysenteriae) was isolated which was multiple drug resistant, exhibiting resistance to chloramphenicol, tetracycline, streptomycin and the sulfonamides. Over the years, and continuing into the present almost every known bacterial pathogen has developed resistance to one or more antibiotics in clinical use. Evidence also began to accumulate that bacteria could pass genes for drug resistance between strains and even between species. For example, antibiotic-resistance genes of staphylococci are carried on plasmids that can be exchanged with Streptococcus and Enterococcus providing the means for acquiring additional genes and gene combinations. Some are carried on transposons, segments of DNA that can exist either in the chromosome or in plasmids. In any case, it is clear that genes for antibiotic resistance can be exchanged between strains and species of bacteria by means of the processes of horizontal gene transmission (HGT).
_
One of the most alarming consequences of antibiotic overuse is the emergence and spread of antibiotic resistant bacteria, which could bring “the end of modern medicine as we know it” (Margaret Chan, Director General of the World Health Organization, 2012). Antibiotic resistance occurs when bacteria lose their sensitivity to antibiotics. It develops when a bacteria mutates or acquires a resistance gene. Resistant bacteria are able to withstand attack by antibiotics, so that standard treatments become ineffective and infections persist and may spread to other people. Resistance of microbial pathogens to antibiotics is increasing worldwide at an accelerating rate, with a concomitant increase in morbidity and mortality associated with infections caused by antibiotic resistant pathogens. At least 2 million people are infected with antibiotic resistant bacteria each year in the US alone, and at least 23,000 people die as a direct result of these infections. In the European Union, an estimated 400,000 patients present with resistant bacterial strains each year, of which 25,000 patients die. Consequently, the World Health Organization has warned that therapeutic coverage will be insufficient within 10 years, putting the world at risk of entering a “post-antibiotic era”, in which antibiotics will no longer be effective against infectious diseases. The Center for Disease Control and Prevention considers this phenomenon “one of the world`s most pressing health problems in the 21st century”.
_
Antimicrobials have been at the forefront in the battle to reduce infectious diseases for much of the past century. They are primarily used to treat infectious diseases in humans and animals, but are also of great value in the prevention of infections when used as prophylaxis, such as in the prevention of infections at the site of a surgical incision or in the prevention of neutropenic sepsis in patients undergoing chemotherapy treatment for cancer. Effective treatment of infection is an essential component of 21st-Century medicine. Modern surgery would be unacceptably dangerous if infections were likely to be untreatable, and cancer chemotherapy and organ transplantation – which suppresses the patient’s immune system – would no longer be viable. As a result, microbes would win a major battle in their long fight with humans. This is now a real risk, as bacteria continue to develop resistance while the flow of new antibiotics has diminished. But while resistance in the targeted organism was a concern, the massive collateral resistance among the myriad “bystander” organisms composing the human microbiome was likely not anticipated.
_
In 1954, the USA produced just under 1 million kilograms of antimicrobials; annual production in this country alone now exceeds 16 million kg. Global consumption of antibiotics in human medicine rose by nearly 40% between 2000 and 2010. Our ability to develop and mass-produce over 25 classes of antimicrobials in seventy years may seem monumental—indeed, many hailed the new antibiotic era as the end of infectious diseases. Unfortunately, the game is in fact rigged. The infinitesimal generation time of a microbe will always confer it the advantage: it has infinitely more opportunities to gain resistance genes than we have to create new antimicrobials. In this race, humans are being outrun: there have been no successful discoveries of new classes of antibiotics since 1987, while new, multi-resistant pathogens such as carbapenem-resistant enterobacteriaceae are spreading with unprecedented alacrity.
_
The figure below shows that antibiotic consumption has markedly increased in developing world as compared to developed world:
_
Spontaneous natural development of antimicrobial resistance in the microorganisms in nature is a slow process. However, the frequent and inappropriate use of a newly discovered antimicrobial drug leads to the development of altered mechanisms in the pathophysiology of the concerned microbes as a survival strategy. Such antibiotic selection pressure kills the susceptible microbes and helps in selective replication of drug resistant bacteria. These resistant bacteria already existed in the population along with the susceptible ones or susceptible bacteria acquired resistance during antimicrobial treatment. Ultimately, such resistant bacteria multiply abundantly and entirely replace the susceptible bacterial population. This results in treatment failure or ineffective management of such infected patients. Antimicrobial resistance has been observed and reported with practically all the newly discovered antimicrobial molecules till date. Antimicrobial resistance makes the treatment of patients difficult, costly and sometimes impossible. Emergence of antimicrobial resistance in pathogens has become a matter of great public health concern. Antimicrobial resistance is well recognised as a global threat to human health. Infections caused by antimicrobial-resistant micro-organisms in hospitals are associated with increased morbidity, mortality and healthcare costs. Resistance has emerged even to newer and more potent antimicrobial agents like carbapenems. Selection and spread of resistant microorganisms in the presence of antimicrobials is facilitated by:
• Irrational use of drugs
• Self-medication
• Misuse of drugs
Antimicrobial resistance is closely linked to inappropriate antimicrobial use. It is estimated that 50% or more of hospital antimicrobial use is inappropriate. There is a need for increased education and awareness about antimicrobial resistance among the public and health-care professionals. One needs to develop and improve the surveillance system for antimicrobial resistance and infectious diseases in general, particularly through improved linkage of data. Nothing will work unless we improve diagnostic testing to ensure more tailored interventions and respond to the opportunities afforded by advances in genomic technologies and point of care testing.
_
Do antibiotics cause microorganisms to become antibiotic resistant?
No. Antibiotics select out the resistant strain. Faulty use of antibiotics or widespread use of antibiotics increases the probability of such selection.
Are antibiotic resistant organisms more virulent?
No. Antibiotic resistant strains appear to be more virulent because we cannot kill them or stop their growth.
_
Since ‘post antibiotic era’ is reported to be “discovery void”; antimicrobial resistance is considered to be the most serious health threats especially for the common infections like sepsis, diarrhea, pneumonia, urinary tract infection, gonorrhea, malaria, tuberculosis, HIV, influenza. Presently, carbapenem resistance is reported worldwide in more than 50% of strains of Klebsiella pneumoniae causing health care associated infections like pneumonia, blood stream infections, infections in the newborn and intensive care units. More than 50% of Escherichia coli strains causing urinary tract infections are reported worldwide to be resistant to fluoroquinolones. Similarly, patients suffering from gonorrhea are reported to be resistant to third generation cephalosporins. High mortality (64%) was seen among patients infected with Methicillin resistant Staphylococcus aureus (MRSA). Over all, the antimicrobial resistance is associated with higher mortality rate, longer hospital stay, delayed recuperation and long term disability. The irony of trend toward progressively more resistant bacteria is that it coincides with a period of dramatically increased understanding of the molecular mechanisms of antimicrobial resistance. Unfortunately, while this insight has resulted in the identification of novel drug targets, it has not yet resulted in effective new chemotherapeutic agents. This paradox stands in sharp contrast to the dramatic progress made in antiviral (notably antiretroviral) therapy in the past ten years, where a number of newly discovered molecular targets have resulted in clinically effective therapeutic agents.
__
Antibiotic resistance has become a major clinical and public health problem within the lifetime of most people living today. Confronted by increasing amounts of antibiotics over the past 60 years, bacteria have responded to the deluge with the propagation of progeny no longer susceptible to them. While it is clear that antibiotics are pivotal in the selection of bacterial resistance, the spread of resistance genes and of resistant bacteria also contributes to the problem. Selection of resistant forms can occur during or after antimicrobial treatment; antibiotic residues can be found in the environment for long periods of time after treatment. Besides antibiotics, there is the mounting use of other agents aimed at destroying bacteria, namely the surface antibacterials now available in many household products. These too enter the environment. The stage is thus set for an altered microbial ecology, not only in terms of resistant versus susceptible bacteria, but also in terms of the kinds of microorganisms surviving in the treated environment. We currently face multi-resistant infectious disease organisms that are difficult and, sometimes, impossible to treat successfully. In order to curb the resistance problem, we must encourage the return of the susceptible commensal flora. They are our best allies in reversing antibiotic resistance.
_
Today we can list a number of organisms in both hospitals and the community that thwart treatment because they are resistant to not one, but to many different antibiotics. The term multidrug resistance (MDR), which initially described resistant mammalian tumour cells, and later strains of Mycobacterium tuberculosis, now describes multidrug resistance in any microorganism—bacterium, fungus or parasite. The emergence of MDR is clearly related to the quantity of antibiotics and how they are being used. It may reflect acquisition of different resistance determinants on the same DNA molecule, or single determinants, such as multidrug pumps, that specify efflux activity against different antibacterials. Besides the known pathogens, the relatively recent appearance of opportunistic organisms, intrinsically resistant to many drugs, is now complicating the advances that we have made in medical technologies. With a larger number of immunocompromised patients and longer time periods spent in an immunocompromised state, these organisms have become ‘specialized’ pathogens—typically attacking only the most vulnerable patients. Among these opportunistic pathogens are the enterococci, the coagulase-negative staphylococci, Pseudomonas aeruginosa and Acinetobacter baumanii. Those physicians attending medical school 20–30 years ago probably did not even discuss these organisms as important pathogens, though today they cause prominent, even potentially lethal, problems in hospitals worldwide. Importantly, organisms known since the early days of clinical microbiology are becoming critical health hazards because of the lack of therapeutic options. A good example, reported by the Public Health Laboratory Service in the UK, is multidrug-resistant Salmonella typhi with resistance to ciprofloxacin, a drug becoming essential in treating this organism. The frequency of resistance to ciprofloxacin was found to be nearly 35%. Some may remember the problems in Central America when resistance to ampicillin and chloramphenicol in S. typhi led to deaths. In these same areas, strains of S. typhi now bear resistance to five or six different agents, including fluoroquinolones. Colistin is the antibiotic that is currently used as a last line of defence. It has rarely been used because it has serious toxic side-effects on the kidneys and most bacteria haven’t developed resistance to colistin because they haven’t been exposed to it. Most but not all. Bacteria have surfaced in meat and animals that carry a gene that makes them resistant to colistin and a woman living in Pennsylvania was found to carry the gene in May 2016. In addition, the CDC has warned that gonorrhea is close to attaining superbug status because AMR has reduced the five treatment options that were available in 2006 to one.
_______
History of antibiotics and antibiotics Resistance:
The management of microbial infections in ancient Egypt, Greece, and China is well-documented. The modern era of antibiotics started with the discovery of penicillin by Sir Alexander Fleming in 1928. Since then, antibiotics have transformed modern medicine and saved millions of lives. Antibiotics were first prescribed to treat serious infections in the 1940s. Penicillin was successful in controlling bacterial infections among World War II soldiers. However, shortly thereafter, penicillin resistance became a substantial clinical problem, so that, by the 1950s, many of the advances of the prior decade were threatened. In response, new beta-lactam antibiotics were discovered, developed, and deployed, restoring confidence. However, the first case of methicillin-resistant Staphylococcus aureus (MRSA) was identified during that same decade, in the United Kingdom in 1962 and in the United States in 1968. Unfortunately, resistance has eventually been seen to nearly all antibiotics that have been developed. Vancomycin was introduced into clinical practice in 1972 for the treatment of methicillin resistance in both S. aureus and coagulase-negative staphylococci. It had been so difficult to induce vancomycin resistance that it was believed unlikely to occur in a clinical setting. However, cases of vancomycin resistance were reported in coagulase-negative staphylococci in 1979 and 1983. From the late 1960s through the early 1980s, the pharmaceutical industry introduced many new antibiotics to solve the resistance problem, but after that the antibiotic pipeline began to dry up and fewer new drugs were introduced. As a result, in 2015, many decades after the first patients were treated with antibiotics, bacterial infections have again become a threat.
_
The figure above shows timeline of antibiotic resistance.
______
Antimicrobial resistance (AMR) is a natural process. It occurs when microorganisms evolve to be able to resist the medicine that has been used to combat them. Resistant microorganisms can survive or even grow in the presence of a concentration of antimicrobial that is usually sufficient to inhibit or kill non-resistant microorganisms of the same species. This important feature was foreseen by Alexander Fleming who, in his speech when receiving the Nobel Prize in Medicine for the discovery of penicillin, issued a warning about the possibility of creating resistant organisms if antibiotics were used irresponsibly. It is likely that AMR began as soon as mass use of antimicrobials by the population began, soon after industrialised production became possible. Antibiotics should only be used when needed as prescribed by health professionals. The prescriber should closely adhere to the five rights of drug administration: the right patient, the right drug, the right dose, the right route, and the right time. Narrow-spectrum antibiotics are preferred over broad-spectrum antibiotics when possible, as effectively and accurately targeting specific organisms is less likely to cause resistance. Cultures should be taken before treatment when indicated and treatment potentially changed based on the susceptibility report. For people who take these medications at home, education about proper use is essential. Health care providers can minimize spread of resistant infections by use of proper sanitation: including hand washing and disinfecting between patients; and should encourage the same of the patient, visitors, and family members. Antibiotic resistance was once confined primarily to hospitals but is becoming increasingly prevalent in family practice settings, making daily therapeutic decisions more challenging. Recent reports of paediatric deaths and illnesses in communities in the United States have raised concerns about the implications and future of antibiotic resistance. Because 20 percent to 50 percent of antibiotic prescriptions in community settings are believed to be unnecessary, primary care physicians must adjust their prescribing behaviors to ensure that the crisis does not worsen. Clinicians should not accommodate patient demands for unnecessary antibiotics and should take steps to educate patients about the prudent use of these drugs. Prescriptions for targeted-spectrum antibiotics, when appropriate, can help preserve the normal susceptible flora. Antimicrobials intended for the treatment of bacterial infections should not be used to manage viral illnesses. Local resistance trends may be used to guide prescribing decisions.
_
Antimicrobial resistance is one of the greatest threats to human health worldwide. It dramatically reduces the probability of effectively treating infections and increases the morbidity and mortality associated with common bacterial diseases. Since the discovery of penicillin in 1928, antimicrobial resistance has been linked to antibiotic use. Recent studies of bacteria in permafrost samples documented the existence of resistance genes 30,000 years ago (D’Costa et al., 2011), emphasizing that antibiotic use and misuse favor resistance through selection pressure (Rolain et al., 2013). It has been demonstrated that antimicrobial resistance prevalence can be diminished through decreased antibiotic consumption (Seppälä et al., 1997). This complex ecological phenomenon depends on individuals, bacterial strains, and mechanisms of resistance (Andersson and Hughes, 2010). Old and new antibiotics vary in their impact on the emergence and spread of resistant bacteria (Sullivan et al., 2001). Bacterial strains resistant to newly developed antibiotics have emerged recurrently (Long and Vester, 2012). Therefore, antimicrobial resistance presents an ongoing challenge that requires a multifaceted approach including (i) biomedical innovation, (ii) improved surveillance of antibiotic consumption and antimicrobial-resistance rates, (iii) prevention of health-care-associated infections and transmission of multidrug-resistant (MDR) bacteria and environmental dissemination, (iv) rapid microbiological diagnosis, and (v) curtailed clinical and veterinary misuse. It is alarming that although bacterial resistance continues to emerge, the rate at which antibiotics are being developed is decreasing. In this context, the reintroduction of previously used antibiotics active against MDR bacteria represents a new alternative for the control of antimicrobial resistance (Pulcini et al., 2012). Indian Health care professionals involved in the treatment of patients with severe infections especially healthcare associated infections will agree that it is commonplace to come across pan-resistant Gram negative bacterial infections where they do not have a single effective antibiotic option. They are, therefore, forced to use a cocktail of antibiotics to which the bacteria is resistant with the infinitesimally small hope of a synergistic effect. Immunocompromised patients especially transplant and cancer chemotherapy patients, who develop infections, are known to have high mortality rates. Until a few years ago we could at least try our powerful antibiotics against these infections. With increasing pan resistant bacteria, we will be forced to stop organ transplantation, bone marrow transplantation and cancer chemotherapy. We are going to face this catastrophic situation in the near future – not in a decade or so but within a few years’ time.
_______
Multidrug tolerance vs. AMR:
Multidrug tolerance or antibiotic tolerance is the ability of a disease-causing microorganism to resist killing by antibiotics or other antimicrobials. It is mechanistically distinct from multidrug resistance: It is not caused by mutant microbes, but rather by microbial cells that exist in a transient, dormant, non-dividing state. Microorganisms that display multidrug tolerance can be bacteria, fungi or parasites. Multidrug tolerance is caused by a small subpopulation of microbial cells termed persisters. Persisters are not mutants, but rather are dormant cells that can survive the antimicrobial treatments that kill the majority of their genetically identical siblings. Persister cells have entered a non- or extremely slow-growing physiological state which makes them insensitive (refractory or tolerant) to the action of antimicrobial drugs. When such persisting microbial cells cannot be eliminated by the immune system, they become a reservoir from which recurrence of infection will develop. Indeed, it appears that persister cells are the main cause for relapsing and chronic infections. Chronic infections can affect people of any age, health, or immune status. Bacterial multidrug or antibiotic tolerance poses medically important challenges. It is largely responsible for the inability to eradicate bacterial infections with antibiotic treatment. Persister cells are highly enriched in biofilms, and it has been suggested that this is the reason that makes biofilm-related diseases so hard to treat. Examples are chronic infections of implanted medical devices such as catheters and artificial joints, urinary tract infections, middle ear infections and fatal lung disease.
Distinction from multidrug resistance:
Unlike resistance, multidrug tolerance is a transient, non-heritable phenotype. Multidrug tolerant persister cells are not antibiotic resistant mutants. Resistance is caused by newly acquired genetic traits (by mutation or horizontal gene transfer) that are heritable and confer the ability to grow at elevated concentrations of antimicrobial drugs. In contrast, multidrug tolerance is caused by a reversible physiological state in a small subpopulation of genetically identical cells, similar to a differentiated cell type. It enables this small subpopulation of microbes to survive the antibiotic killing of their surrounding siblings. Persisting cells resume growth when the antimicrobial agent is removed, and their progeny is sensitive to antimicrobial agents.
Molecular mechanisms:
The molecular mechanisms that underlie persister cell formation and multidrug tolerance are largely unknown. Persister cells are thought to arise spontaneously in a growing microbial population by a stochastic genetic switch, although inducible mechanisms of persister cell formation have been described. Owing to their transient nature and relatively low abundance, it is hard to isolate persister cells in sufficient numbers for experimental characterization, and only a few relevant genes have been identified to date. The best-understood persistence factor is the E. coli high persistence gene, commonly abbreviated as hipA. Although tolerance is widely considered a passive state, there is evidence indicating it can be an energy-dependent process. Persister cells in E. coli can transport intracellular accumulations antibiotic using an energy requiring efflux pump called TolC.
Potential treatment:
In May 2011, it was reported that the addition of certain metabolites can help suppress multidrug tolerance in numerous species of bacteria, including E. coli and S. aureus, by “the generation of a proton-motive force which facilitates aminoglycoside uptake”. Phage therapy, where applicable, entirely circumvents antibiotic resistance.
____
____
The ecology of antibiotic resistance:
The impact of the drug selection process can be largely confined to the individual taking the antibiotic if widespread antibiotic usage is absent. After therapy, the selected resistant commensal strains will eventually be ‘diluted out’ and their growth will be suppressed by the return of drug-susceptible, natural competitors. If, however, whole populations are being treated with the same class of antibiotic, susceptible strains will have little opportunity to recolonize their niche and resistant strains will acquire an important advantage. The resulting ecological imbalance produces a potentially serious environmental pool of resistance genes. Ecologically speaking, it is the density of antibiotic usage that enhances resistance selection and its effects. The ‘selection density’ involves the total amount of antibiotic being applied to a geographically defined number of individuals in a setting, whether it is the home, daycare center, hospital or farm. Each individual becomes a ‘factory’ of resistant bacteria that enter the environment. The disparity between resistance rates in the local community and those in city hospitals reflects differential ecological effects of antibiotic use. The end result of the selective pressure will reflect the number of individuals who are contributing resistant bacteria to that environment and the residual number of surviving, susceptible bacteria. The ecological effects of antibiotics make them unique therapeutic agents. They are ‘societal drugs’ in which individual use affects others sharing that environment. For example, antibiotic treatment for acne was found to produce an MDR skin flora not only in the individual with acne, but also in other members of the household. High numbers of MDR bacteria were found in the intestinal flora of ambulatory individuals in the Boston area, even though none had recently taken an antibiotic. In Nepal, resistance rates in individuals were found to correlate more with the total community use of antibiotics than with the individual’s own use. In addition, the selection of resistance continues because antimicrobials persist, largely intact, in natural environments. Antimicrobials in waste waters are being reported with increasing frequency and are potentially important contributors to the environmental selection of antibiotic-resistant organisms. The findings suggest that one approach to the antibiotic resistance problem could be to design drugs that self-destruct after treatment, thereby removing a contributing factor in the propagation of resistance.
____
____
UDR vs. MDR:
All future clinical trials need to be designed with antimicrobial resistance in mind. Wild-type bacteria – which are susceptible to most antibiotics – have become rare, while those that developed usual drug resistance (UDR) are more common. One of the most frightening prospects is that UDR bacteria have the potential to evolve into multiple-drug resistant (MDS) bacteria or microorganisms with extensive multi-drug resistance. Driving an organism back into the less dangerous UDR category requires new antimicrobials, while the development of new antibiotics is more challenging in the context of widespread antimicrobial resistance. New clinical trial designs and strategies need to be devised and made to work.
_
Multiple drug resistant (MDR) organisms:
Multiple drug resistance (MDR), multidrug resistance or multiresistance is antimicrobial resistance shown by a species of microorganism to multiple antimicrobial drugs. The types most threatening to public health are MDR bacteria that resist multiple antibiotics; other types include MDR viruses, fungi, and parasites (resistant to multiple antifungal, antiviral, and antiparasitic drugs of a wide chemical variety). Recognizing different degrees of MDR, the terms extensively drug resistant (XDR) and pandrug-resistant (PDR) have been introduced. The definitions were published in 2011 in the journal Clinical Microbiology and Infection and are openly accessible. Multiple drug resistant organisms are resistant to treatment with several, often unrelated, antimicrobial agents.
Some of the most important types of multiple drug resistant organisms that have been encountered include:
1. MRSA – methicillin/oxacillin-resistant Staphylococcus aureus
2. VRE – vancomycin-resistant enterococci
3. ESBLs – extended-spectrum beta-lactamases producing Gram-negative bacteria (which are resistant to cephalosporins and monobactams)
4. PRSP – penicillin-resistant Streptococcus pneumoniae
MRSA and VRE are the most commonly encountered multiple drug resistant organisms in patients residing in non-hospital healthcare facilities, such as nursing homes and other long-term care facilities. PRSP are more common in patients seeking care in outpatient settings such as physicians’ offices and clinics, especially in paediatric settings. ESBLs are most often encountered in the hospital (intensive care) setting, but MRSA and VRE also have a significant nosocomial ecology.
5. Carbapenemase producing Klebsiella pneumonia (KPC)
6. MultiDrug-Resistant gram negative rods (MDR GNR) MDRGN bacteria such as Enterobacter species, E.coli, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa.
7. A group of gram-positive and gram-negative bacteria of particular recent importance have been dubbed as the ESKAPE group (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species).
8. Multi-drug-resistant tuberculosis
_
One example of MDR bacteria:
Blood, sputum, and pleural fluid cultures taken from our nurse-patient in the ICU grew Acinetobacter baumannii, a known multidrug-resistant gram-negative bacillus. Upon this finding, her antibiotics were changed, initially to imipenem-cilastatin and tigecycline, and when susceptibility data were available, to amikacin. She was on antibiotics for 3 weeks. Her hospital course, however, remained stormy and was complicated by pleural decortication, hypotension, and renal failure which required haemodialysis. After 2 months, she was discharged from the ICU to a rehabilitation facility, where she remained for an additional month.
_
Antifungal MDR:
Yeasts such as Candida species can become resistant under long term treatment with azole preparations, requiring treatment with a different drug class. Scedosporium prolificans infections are almost uniformly fatal because of their resistance to multiple antifungal agents.
Antiviral MDR:
HIV is the prime example of MDR against antivirals, as it mutates rapidly under monotherapy. Influenza virus has become increasingly MDR; first to amantadine, then to neuraminidase inhibitors such as oseltamivir, (2008-2009: 98.5% of Influenza A tested resistant), also Cytomegalovirus can become resistant to ganciclovir and foscarnet under treatment, especially in immunosuppressed patients. Herpes simplex virus rarely becomes resistant to acyclovir preparations, mostly in the form of cross-resistance to famciclovir and valacyclovir, usually in immunosuppressed patients.
Antiparasitic MDR:
The prime example for MDR against antiparasitic drugs is malaria. Plasmodium vivax has become chloroquine and sulfadoxine-pyrimethamine resistant a few decades ago, and as of 2012 artemisinin-resistant Plasmodium falciparum has emerged in western Cambodia and western Thailand. Toxoplasma gondii can also become resistant to artemisinin, as well as atovaquone and sulfadiazine, but is not usually MDR. Antihelminthic resistance is mainly reported in the veterinary literature, for example in connection with the practice of livestock drenching and has been recent focus of FDA regulation.
_
Superbug:
Many of the bacterial pathogens associated with epidemics of human disease have evolved into multidrug-resistant (MDR) forms subsequent to antibiotic use. For example, MDR M. tuberculosis is a major pathogen found in both developing and industrialized nations and became the 21th-century version of an old pathogen. Other serious infections include nosocomial (hospital-linked) infections with Acinetobacter baumannii, Burkholderia cepacia, Campylobacter jejuni, Citrobacter freundii, Clostridium difficile, Enterobacter spp., Enterococcus faecium, Enterococcus faecalis, Escherichia coli, Haemophilus influenzae, Klebsiella pneumoniae, Proteus mirabilis, Pseudomonas aeruginosa, Salmonella spp., Serratia spp., Staphylococcus aureus, Staphylococcus epidermidis, Stenotrophomonas maltophilia, and Streptococcus pneumoniae. The term “superbugs” refers to microbes with enhanced morbidity and mortality due to multiple mutations endowing high levels of resistance to the antibiotic classes specifically recommended for their treatment; the therapeutic options for these microbes are reduced, and periods of hospital care are extended and more costly. In some cases, superresistant strains have also acquired increased virulence and enhanced transmissibility. Realistically, antibiotic resistance can be considered a virulence factor.
_____
_____
Global concern of AMR:
_
Why AMR a global concern?
New resistance mechanisms emerge and spread globally threatening our ability to treat common infectious diseases, resulting in death and disability of individuals who until recently could continue a normal course of life. Without effective anti-infective treatment, many standard medical treatments will fail or turn into very high risk procedures.
1. AMR kills: Infections caused by resistant microorganisms often fail to respond to the standard treatment, resulting in prolonged illness, higher health care expenditures, and a greater risk of death. As an example, the death rate for patients with serious infections caused by common bacteria treated in hospitals can be about twice that of patients with infections caused by the same non-resistant bacteria. For example, people with MRSA (methicillin-resistant Staphylococcus aureus, another common source of severe infections in the community and in hospitals) are estimated to be 64% more likely to die than people with a non-resistant form of the infection.
2. AMR hampers the control of infectious diseases: AMR reduces the effectiveness of treatment; thus patients remain infectious for a longer time, increasing the risk of spreading resistant microorganisms to others. For example, the emergence of Plasmodium falciparum resistance to artemisinin in the Greater Mekong subregion is an urgent public health concern that is threatening global efforts to reduce the burden of malaria. Although MDR-TB is a growing concern, it is still largely under-reported, compromising control efforts.
3. AMR increases the costs of health care: When infections become resistant to first-line drugs, more expensive therapies must be used. A longer duration of illness and treatment, often in hospitals, increases health care costs as well as the economic burden on families and societies.
4. AMR jeopardizes health care gains to society: The achievements of modern medicine are put at risk by AMR. Without effective antimicrobials for prevention and treatment of infections, the success of organ transplantation, cancer chemotherapy and major surgery would be compromised.
5. AMR has the potential to threaten health security, and damage trade and economies: The growth of global trade and travel allows resistant microorganisms to be spread rapidly to distant countries and continents through humans and food. Estimates show that AMR may give rise to losses in Gross Domestic Product of more than 1% and that the indirect costs affecting society may be more than 3 times the direct health care expenditures. It affects developing economies proportionally more than developed ones.
_
The figure below shows rise of AMR worldwide:
_
Global action plans:
1. AMR was the focus of the World Health Organisation (WHO) World Health Day 2011, with all countries urged to commit to a comprehensive plan to tackle the issue, including reducing the use of antibiotics in food producing animals, enhanced infection prevention and control and innovations for new research.
_
2. In 2013 WHO set up a Strategic and Technical Advisory Group on antimicrobial resistance to review and help shape a global strategy to tackle the growing challenge of antimicrobial resistance (AMR), and to advise WHO on the coordination role it should be playing in the fight against AMR.
_
3. In April 2014 the WHO published Antimicrobial resistance: global report on surveillance. This report released April 2014 stated, “This serious threat is no longer a prediction for the future, it is happening right now in every region of the world and has the potential to affect anyone, of any age, in any country. Antibiotic resistance—when bacteria change so antibiotics no longer work in people who need them to treat infections—is now a major threat to public health.” The report notes that resistance is occurring across many different infectious agents but the report focuses on antibiotic resistance in seven different bacteria responsible for common, serious diseases such as bloodstream infections (sepsis), diarrhoea, pneumonia, urinary tract infections and gonorrhoea. The results are cause for high concern, documenting resistance to antibiotics, especially “last resort” antibiotics, in all regions of the world.
Key findings from the report include:
•Resistance to the treatment of last resort (carbapenem antibiotics) for life-threatening infections caused by a common intestinal bacteria, Klebsiella pneumonia has spread to all regions of the world. K. pneumoniae is a major cause of hospital-acquired infections such as pneumonia, bloodstream infections, infections in newborns and intensive-care unit patients. In some countries, because of resistance, carbapenem antibiotics would not work in more than half of people treated for K. pneumoniae infections.
•Resistance to one of the most widely used antibacterial medicines for the treatment of urinary tract infections caused by E. coli–fluoroquinolones–is very widespread. In the 1980s, when these drugs were first introduced, resistance was virtually zero. Today, there are countries in many parts of the world where this treatment is now ineffective in more than half of patients.
•Treatment failure to the last resort of treatment for gonorrhoea–third generation cephalosporins–has been confirmed in Austria, Australia, Canada, France, Japan, Norway, Slovenia, South Africa, Sweden and the United Kingdom. An estimated 106 million people are infected with gonorrhoea every year (2008 estimates).
•Antibiotic resistance causes people to be sick for longer and increases the risk of death. For example, people with MRSA (methicillin-resistant Staphylococcus aureus) are estimated to be 64% more likely to die than people with a non-resistant form of the infection. Resistance also increases the cost of health care with lengthier stays in hospital and more intensive care required.
• There are significant gaps in surveillance, and a lack of standards for methodology, data sharing and coordination.
Key findings from AMR surveillance in disease-specific programmes are as follows:
a) Although multidrug-resistant TB is a growing concern, it is largely under-reported, compromising control efforts.
b) Foci of artemisinin resistance in malaria have been identified in a few countries. Further spread, or emergence in other regions, of artemisinin resistant strains could jeopardize important recent gains in malaria control.
c) Increasing levels of transmitted anti-HIV drug resistance have been detected among patients starting antiretroviral treatment.
_
The report reveals that key tool to tackle antibiotic resistance–such as basic systems to track and monitor the problem–show gaps or do not exist in many countries. While some countries have taken important steps in addressing the problem, every country and individual needs to do more. Other important actions include preventing infections from happening in the first place–through better hygiene, access to clean water, infection control in health-care facilities, and vaccination–to reduce the need for antibiotics. WHO is also calling attention to the need to develop new diagnostics, antibiotics and other tools to allow healthcare professionals to stay ahead of emerging resistance. This report is kick-starting a global effort led by WHO to address drug resistance. This will involve the development of tools and standards and improved collaboration around the world to track drug resistance, measure its health and economic impacts, and design targeted solutions.
_
4. UN meeting tackles the ‘fundamental threat’ of antibiotic-resistant superbugs in on September 21, 2016:
All 193 UN member states have agreed to combat the proliferation of drug-resistant infections, estimated to kill more than 700,000 people each year. The UN secretary general, Ban Ki-moon, said antimicrobial resistance is a “fundamental threat” to global health and safety at the first general assembly meeting on drug-resistant bacteria. It is only the fourth time the general assembly has held a high-level meeting for a health issue. “If we fail to address this problem quickly and comprehensively, antimicrobial resistance will make providing high-quality universal healthcare coverage more difficult if not impossible,” said Ban. “It will undermine sustainable food production. And it will put the sustainable development goals in jeopardy.” Just before world leaders convened for the meeting, all 193 member states agreed in a declaration signed on Wednesday to combat the proliferation of antibiotic resistance. “Antimicrobial resistance poses a fundamental threat to human health, development, and security. The commitments made today must now be translated into swift, effective, lifesaving actions across the human, animal, and environmental health sectors. We are running out of time,” said Dr Margaret Chan, Director-General of WHO.
World leaders reaffirmed their commitment to:
•strengthen regulation of antimicrobials,
•improve knowledge and awareness,
•promote best practices
•foster innovative approaches using alternatives to antimicrobials and new technologies for diagnosis and vaccines
_____
European Antibiotic Awareness Day (EAAD):
Every year, European Antibiotic Awareness Day is held on November 18. It’s a European-wide public health initiative that encourages the responsible use of antibiotics.
_
World Antibiotic Awareness Week 2016: “Antibiotics: Handle with care”:
This year World Antibiotic Awareness Week is held from 14 to 20 November 2016. World Antibiotic Awareness Week aims to increase awareness of global antibiotic resistance and to encourage best practices among the general public, health workers and policy makers to avoid the further emergence and spread of antibiotic resistance. The theme of the campaign, Antibiotics: Handle with Care, reflects the overarching message that antibiotics are a precious resource and should be preserved. They should be used to treat bacterial infections, only when prescribed by a certified human or animal health professional. Antibiotics should never be shared or saved for the future. WHO is encouraging all Member States, health partners and students, and the public to join this campaign and help raise awareness of antibiotic resistance.
____
From an epidemiological and methodological standpoint the comparison of antimicrobial resistance from different countries is very difficult. Reasons for these difficulties are that:
1. Different antimicrobial agents are tested.
2. Different systems for antimicrobial susceptibility testing are used.
3. Different breakpoints for antimicrobial susceptibility are used.
4. Data from point prevalence studies are used for longitudinal comparisons; e.g. studies on antibiotic resistance performed in 1970 and 1990 are compared, in spite of differences in study conditions and methodology.
5. Only the resistant strains are tested.
6. Differences between the prevalence of resistant strains from local practices and university hospitals are not taken into account.
____
Antibiotic resistance highest in poorer countries, research shows:
The research conducted by the Centre for Disease Dynamics, Economics & Policy (CDDEP) finds a strong correlation between a country’s income level and antimicrobial resistance. This study reveals that countries with low national income have higher levels of antimicrobial resistance. The study conducted by CDDEP researchers highlights an urgent need for countries with low-income level to address the prevalence of AMR. The findings show that AMR levels are highest in the poorest countries. Poor environmental sanitation, poor infection control practices and lack of stewardship programs in healthcare facilities in low-income countries are the major contributors. The burden of bacteria is higher in the lower and middle income countries due to lack of sanitation conditions. This further results in the higher prevalence of AMR. The study assessed the association between the gross national income per capita (GNIPC) of a country and level of AMR of three common pathogens causing infections in hospitals and the community. The research was conducted for 2013-14, across 45 countries divided into high, upper-middle and lower middle income economies. The model predicted AMR prevalence of 12% in high-income countries, 31% in upper-middle income countries and 78% in lower-middle income countries. The study points that resistance to antimicrobials leads to huge health costs as well as higher rates of mortality. This is because effective antibiotics with AMR are more expensive and thus, unaffordable by a major proportion of people living in their resource limited environment. It also leads to higher resistance to last resort antibiotics. The study cites various reports and papers explaining reasons for transmission of antimicrobial resistance. It mainly occurs from contact with animals, other human beings and the environment. This is facilitated by factors which are prevalent in the low and middle income countries including high population density, lack of access to clean water, suboptimal sewage systems, poor sanitation and poor healthcare infection control practices. Lack of regulation on antimicrobial use in farming and pharmaceutical industry pollution are other important reasons identified by the study.
____
____
Global spread of AMR:
If you travel internationally, you can pick up an infection not commonly found at home through contact with:
•people
•food
•water
•animals
•contaminated surfaces
International travel may include trips for business, pleasure or even to have a medical procedure. With the growth of global trade and travel, resistant microorganisms can spread promptly to any part of the world.
_
The ability of influenza virus to spread globally has long been recognized, with several pandemics having been recorded over the last 100 years. The pandemic spread of this infectious agent is due not only to person-to-person spread in local environments but also to the mobility of human populations facilitated by the ready availability of air and ground transportation systems. Individuals incubating an infection may travel between countries or even continents in a matter of hours or days, after which they become infectious, thus transmitting the infection over vast distances. However, there is increasing appreciation that influenza virus is not unique and that many other pathogens are also transmitted internationally, including bacteria that are resistant to antibiotics. The global dissemination of antibiotic-resistant bacteria has received much attention, particularly over the last 25 years, following reports of the international spread of multi-resistant Streptococcus pneumoniae (Mun˜oz et al., 1991), meticillin-resistant Staphylococcus aureus (Johnson, 2011; Stefani et al., 2012) and resistant Enterobacteriaceae, particularly strains resistant to cephalosporins due to the production of CTX-M type extended-spectrum b-lactamases and strains producing carbapenemases such as KPC (van der Bij & Pitout, 2012). The rapidity with which new types of antibiotic resistance can disseminate globally following their initial emergence or recognition is exemplified by the novel carbapenemase New Delhi metallo-blactamase (NDM). The first documented case of infection caused by bacteria producing NDM occurred in 2008, although retrospective analyses of stored cultures have identified the gene encoding this enzyme (blaNDM) in Enterobacteriaceae isolated in 2006. Since its first description, NDM carbapenemase has been reported from 40 countries worldwide, encompassing all continents except South America and Antarctica.
_
_
Antimicrobial resistance is a global health challenge:
Antimicrobial resistance is a global threat that spans all countries, even those with lower consumption of AMTs. The epidemiology of resistance is multinational and there is consolidated evidence that resistant microorganisms do not recognize boundaries. Patients as well as medical personnel or even healthy people may bring ARMs to other hospitals, cities or countries. For example, the first strain of a methicillin-resistant variant of S. aureus (MRSA) was isolated in the United Kingdom 2 year after the introduction of methicillin in 1959. During the 1960s variants of this strain were isolated in many European countries and, then, during the 1970s in other parts of the worlds including Australia, Japan and the United States. MRSA is now a major cause of nosocomial infections worldwide (Deurenberg et al., 2007). Increased mobility and globalization are reducing the time needed for antibiotic-resistant microorganisms to spread. So, if half a century ago MRSA took about two decades to spread to Europe and, then to the rest of the world, a carbapenem-resistant strain of Klebsiella needed only 5 years to spread from the United States, where it was identified in 2003, to Israel (2005) to the United Kingdom, Italy and Colombia (2008) (McKenna, 2013).
__
_____
_____
Threat of AMR:
_
AMR statistics and epidemiology:
The most recent worldwide estimates of global antibiotic resistance, published by the World Health Organisation (WHO) in 2014, list Klebsiella pneumoniae, Escherichia coli and Staphylococcus aureus as the three agents of greatest concern, associated with both hospital- and community-acquired infections. In 2014 in India, 57% of the infections caused by Klebsiella pneumoniae, a dangerous superbug found in hospitals, were found to be resistant to the last-resort antibiotic class of drugs carbapenems, up from 29% in 2008. It was more than 60% resistant for four out of five drug classes tested. This is a dangerous trend as the Klebsiella bug is around 80% resistant to the drug class 3rd generation cephalosporins, 73% resistant to fluoroquinolones and 63% to aminoglycosides. For comparison, these drugs are still effective against Klebsiella infections in 90% of cases in the US and over 95% of cases in Europe. India also has the highest rates of Escherichia coli (E. coli) resistance, with strains of E. coli being more than 80% resistant to three different drug classes, thus increasingly limiting treatment options. E. coli resistance is also high and rising for many drug types in other regions too. In Europe, North America, South and Southeast Asia and parts of Africa, resistance to aminopenicillins – a broad-spectrum antibiotic class – is around 50%. Incidence of methicillin-resistant Staphylococcus aureus (MRSA), a highly dangerous type of resistant pathogen contracted mostly in hospitals, has declined in Europe, the US, Canada and South Africa during the past eight years. However, it is rising in sub-Saharan Africa, India, Latin America and Australia and was recorded at 47% in India in 2014 and 90% in Latin American hospitals in 2013. In Nigeria, some studies suggest that as many as 88% of Staphylococcus aureus infections cannot be treated with methicillin — once a potent weapon against the microbe.
_
Antibiotic-resistant infections are already widespread in the U.S. and across the globe. A 2011 national survey of infectious-disease specialists, conducted by the IDSA Emerging Infections Network, found that more than 60% of participants had seen a pan-resistant, untreatable bacterial infection within the prior year. Many public health organizations have described the rapid emergence of resistant bacteria as a “crisis” or “nightmare scenario” that could have “catastrophic consequences.” The CDC declared in 2013 that the human race is now in the “post-antibiotic era,” and in 2014, the World Health Organization (WHO) warned that the antibiotic resistance crisis is becoming dire. MDR bacteria have been declared a substantial threat to U.S. public health and national security by the IDSA and the Institute of Medicine, as well as the federal Interagency Task Force on Antimicrobial Resistance. Among gram-positive pathogens, a global pandemic of resistant S. aureus and Enterococcus species currently poses the biggest threat. MRSA kills more Americans each year than HIV/AIDS, Parkinson’s disease, emphysema, and homicide combined. Vancomycin-resistant enterococci (VRE) and a growing number of additional pathogens are developing resistance to many common antibiotics. The global spread of drug resistance among common respiratory pathogens, including Streptococcus pneumoniae and Mycobacterium tuberculosis, is epidemic. Gram-negative pathogens are particularly worrisome because they are becoming resistant to nearly all the antibiotic drug options available, creating situations reminiscent of the pre-antibiotic era. The emergence of MDR (and increasingly pan-resistant) gram-negative bacilli has affected practice in every field of medicine. The most serious gram-negative infections occur in health care settings and are most commonly caused by Enterobacteriaceae (mostly Klebsiella pneumoniae), Pseudomonas aeruginosa, and Acinetobacter. MDR gram-negative pathogens are also becoming increasingly prevalent in the community. These include extended-spectrum beta-lactamase-producing Escherichia coli and Neisseria gonorrhoeae.
_
CDC assessment of AMR threats:
The CDC assessed antibiotic-resistant bacterial infections according to seven factors: clinical impact, economic impact, incidence, 10-year projection of incidence, transmissibility, availability of effective antibiotics, and barriers to prevention. The threat level of each bacteria was then classified as “urgent,” “serious,” or “concerning” as seen in the figure below. In general, threats that are urgent or serious require more monitoring and prevention activities, whereas those considered concerning require less.
__
Antibiotics have revolutionized medicine in many respects, and countless lives have been saved; their discovery was a turning point in human history. Regrettably, the use of these wonder drugs has been accompanied by the rapid appearance of resistant strains. Medical pundits are now warning of a return to the preantibiotic era; a recent database lists the existence of more than 20,000 potential resistance genes (r genes) of nearly 400 different types, predicted from available bacterial genome sequences. Fortunately, the number existing as functional resistance determinants in pathogens is much smaller. Some experts have warned that AMR threatens to take us back to the pre-antibiotic era, but actually, it’s worse than that. This will be a post-antibiotic era, where the germs are stronger and more resilient and replacement antibiotics are rare. Imagine an infected cut that becomes life threatening, or pneumonia once again becoming a mass killer. Many of the medical advances we currently take for granted such as cancer treatment, transplantation, surgery and neonatal care―all of which rely on antibiotics to control infection―would be impossible. Nowadays, about 70 percent of the bacteria that cause infections in hospitals are resistant to at least one of the drugs most commonly used for treatment. Some organisms are resistant to all approved antibiotics and can only be treated with experimental and potentially toxic drugs. An alarming increase in resistance of bacteria that cause community acquired infections has also been documented, especially in the staphylococci and pneumococci (Streptococcus pneumoniae), which are prevalent causes of disease and mortality. In a recent study, 25% of bacterial pneumonia cases were shown to be resistant to penicillin, and an additional 25% of cases were resistant to more than one antibiotic.
_
Risks to human health:
Antibiotic resistance is a growing public health concern worldwide. When a person is infected with an antibiotic-resistant bacterium, not only is treatment of that patient more difficult, but the antibiotic-resistant bacterium may spread to other people. The number of bacteria that are resistant to antibiotics is increasing. The danger of antibiotic resistance is that treatable illnesses, such as pneumonia, tuberculosis, or minor infections could become incurable. This would put a greater economic and emotional burden on families and on our healthcare system. Antibiotic resistance results in a decreased ability to treat infections and illnesses in people, animals and plants. This can lead to the following problems:
•increased human illness, suffering and death,
•increased cost and length of treatments, and
•increased side effects from the use of multiple and more powerful medications.
_
Patients infected by ARMs are significantly more likely to develop complications and long-term sequelae. Once an infective microorganism enters a body, it can move from the primary site of infection to other sites. ARMs can usually benefit from extra time to multiply, spread to other organs and to develop complications. For example, patients infected by the methicillin resistant (MRSA) strain of S. aureus have a risk of developing any complication which is 69% higher compared to similar patients infected by its methicillin-susceptible variant (MSSA) as seen in the figure below. The single most frequent complication is a progression of the local infection [relative risk (RR) equal to 3.25 – i.e. a persons infected by MRSA is 3.25 times more likely to have a local progression of the infection compared to a person infected by MSSA]. If the infective microorganism enters the circulatory system and spreads widely, it may cause sepsis (i.e. whole-body inflammatory response to an infection) and, eventually, a shock. Patients infected by ARMs are more likely to develop sepsis and 12% more likely to develop shock. Some other serious long-term complications that are more likely to develop include sequelae in the central nervous system (RR 1.7) or limb loss (RR 1.13).
_
Who is most at risk?
In general, certain groups of people have an increased risk for getting infections. This means they are also at an increased risk of antibiotic resistance.
At risk groups include:
•infants, especially premature babies, as they may not have strong immune systems;
•seniors, particularly those living in long-term care facilities or seniors’ residences. This is because they:
-may be exposed to more infections than the average person,
-are in close contact with many others, and
-may have a weakened immune system due to illness or extended antibiotic use.
•people who are homeless or living in crowded or unhygienic conditions where it is easy to contract infections, and
•people with weakened immune systems due to illness or injury.
_
At risk groups based on behaviours and settings include:
•people in healthcare facilities and day care centres, or other settings where infections can easily spread, especially if infection prevention and control measures are not followed,
•people who do not practice good infection prevention and control behaviours like hand hygiene,
•people who do not store, handle or prepare food safely,
•people who have occupations that put them at a greater risk for exposure to bacteria or infectious diseases such as: physicians, nurses, veterinarians, slaughter house and meat processing plants workers, and farmers.
____
Consequences of antibiotic resistance:
_____
Antimicrobial resistant infections place a great burden on healthcare systems, veterinary practices and society in general, with wide-ranging impact from farms to public health:
•Treatment options are unavailable or limited; available antibiotics may be less effective, more expensive and sometimes harmful to patients
•Resistance means higher mortality risk, longer hospital stays, delayed recuperation, and sometimes long-term disability for patients
•Drug resistant organisms are behind many outbreaks of healthcare associated infection
•Rising resistance in humans has led to measures to restrict the use of certain antibiotics in the veterinary field, limiting therapeutic options.
•While the problem of antimicrobial resistant organisms is most acute in the healthcare setting, there is strong evidence that transmission is increasing in the community
•In addition, antimicrobial resistance is a serious problem in the field of livestock animal production and even in companion animals.
_____
Clinical and economic burden of AMR:
_
AMR mortality:
__
The duration of hospital stays for patients with antibiotic-resistant infections was found to be prolonged by 6.4 to 12.7 days, collectively adding an extra eight million hospital days. Estimates regarding the medical cost per patient with an antibiotic-resistant infection range from $18,588 to $29,069. The Centers for Disease Control and Prevention (CDC) estimates that 2 million illnesses and 23,000 deaths are caused by drug-resistant bacteria in the United States each year. The annual impact of antibiotic resistant infections on U.S. economy is estimated to be $20 – $35 billion with an additional cost to society for lost productivity of $35 billion / year. This results in 8 million additional days in hospitals each year, higher hospital cost and re-admissions. In Europe, AMR is associated with approximately 25 000 deaths annually. A report commissioned by the British government in 2013 estimated that global deaths from AMR are currently 700,000, but that by 2050 AMR could be responsible for 10 million deaths per year and at least $100 trillion (more than the size of the current world economy) in lost output. Both these figures are conservative, because the report only looked at the impact of six antibiotic-resistant pathogens, and did not include the costs of healthcare or secondary social costs. While AMR can affect anyone, in any country, the burden of antimicrobial resistance is not evenly distributed around the globe. Poorer countries and those with restricted access to modern health care feel the burden of AMR more fully than their more developed neighbours. For example, if the present situation continues, it is estimated that by 2050 one-quarter of all deaths in Nigeria could be caused by AMR. This is scary data. AMR is a far more serious threat than conditions such as Zika and Ebola, which receive extensive media coverage. If we fail to tackle this problem we may soon be in the post-antibiotic era, where simple infections will be able to kill because we won’t have antibiotics to treat them. The gains we have made in modern medicine will vanish.
_
__
Antimicrobial resistance—a threat to neonate survival:
The DeNIS study (Delhi Neonatal Infection) followed a group of 88,636 newborn infants for 3 years starting July 2011. The doctors tracked babies born in three of Delhi’s largest hospitals- AIIMS, Safdarjung Hospital and Maulana Azad Medical College -as they were subsequently admitted to the Intensive Care Units. Out of over 88,000 children, 13,530 were ‘enrolled’ in the study – that is, admitted to the ICU. Three ‘superbugs’ in particular – Klebsiella, Acinetobacter, and E. coli – were associated with more than half (53 per cent) of the infections. Out of this 1,934 babies (14 per cent) were resistant to drugs and 496 babies (26 per cent) died due to causes attributable to drug resistance and formerly curable infections. Multi drug resistance was the highest with Acinetobacter in 181 cases, a staggering 82 per cent. Resistance to Klebsiella was found in 54 per cent and E. coli in 38 per cent. Estimates indicate that 56,524 neonates die each year from neonatal sepsis caused by bacteria resistant to first-line antibiotics in India; the toll in Pakistan is 25,692 neonates. In Tanzania, 40% of the 300 neonates with sepsis at a neonatal unit tracked in one study had early onset sepsis and 47% had a positive blood culture. Of those, 29% of neonates who were culture positive died compared with 9% who were culture negative. Mortality was increased in neonates with a Gram-negative bacterial infection, extended-spectrum β lactamase producing organism or methicillin-resistant Staphylococcus aureus. Many neonates in hospitals in south Asia are now treated with carbapenems as a first-line therapy for sepsis or presumed sepsis. Against this backdrop, the widespread availability and antimicrobial use in community settings and the contribution of antimicrobial resistance as a complicating factor in neonate sepsis becomes extremely important. Notwithstanding the importance of preventive strategies to reduce the risks and burden of neonatal infections, early detection and prescribing of appropriate antibiotics will remain the cornerstone of management strategies. The DeNIS study highlights the serious risk associated with neonatal sepsis and resistance in health-care facilities that would rank among the better performing hospitals in a large middle-income country. With an increased focus on institutionalising births in India and other low-income and middle-income countries, the quality of care and infection control in health-care institutions must receive greater attention and resources.
_
Limitations of clinical and economic impact of AMR:
There are few reliable studies of the effect on patient outcomes following an infection with an AMR pathogen. Estimating deaths attributable to AMR is problematic. Patients most susceptible to AMR infections are often those with co-morbidities. Whether the death was directly attributable to an AMR bacterial infection or other co-existing conditions, complicated by the AMR, is often unclear. Determining the true economic impact of antimicrobial drug resistance is a challenge because so many variables and perspectives are involved. Better methods are needed to assess the practical implications for those from all perspectives, whether prescriber, patient, health-care business, pharmaceutical company, or the public. Because studies completed to date have been hampered by their small size and lack of uniformity, validity of the information provided is unclear and extrapolating the studies to regional or national or international levels is questionable. Many studies that attempt to estimate the clinical and economic impact of healthcare infections (including those associated with AMR) fail to account appropriately for length of stay and risk of death. This is an extremely important methodological issue that can severely bias estimation of these key economic drivers. Blot et al found that the data on the impact of drug resistance in nosocomial infections were conflicting and depended on the way confounding variables were accounted for.
_____
_____
Is India epicentre of AMR?
Lord Jim O’Neill, the renowned economist who coined the acronym BRICS and predicted that India would become the second or the third largest economy by 2050, warned that India will fail to reach this “exciting potential” if it doesn’t scale up its effort to tackle the rising antimicrobial resistance. Speaking following the launch of a paper on ‘Infection Prevention, Control and Surveillance’ by the Review on Antimicrobial Resistance that he chairs, he said by 2050, antimicrobial resistance would claim an estimated million lives in India. The productivity of the Indian economy will be severely impaired. India will never reap the demographic dividend, which would have been a global advantage for the country. He described India’s antibiotic resistance problem as complicated. For starters, he said there is no accurate data on resistance. And then, India has a billion people and a lot of them live in poverty and need better access to health care. Antibiotic resistance is a global public health threat, but nowhere is it as stark as in India. The crude infectious disease mortality rate in India today is 416.75 per 100,000 persons and is twice the rate prevailing in the United States when antibiotics were introduced (roughly 200 per 100,000 persons). A mix of poor public health systems and hospital infection, high rates of infectious disease, inexpensive antibiotics, and rising incomes is coming together to increase prevalence of resistant pathogens and is increasing the burden of untreatable neonatal sepsis and health-care-associated infections.
_
Antibiotic Use in India:
_
Antibiotic use is a major driver of resistance. Last year, a study published by researchers from Princeton University, which analysed global trends of antibiotic consumption between 2000 and 2010, discovered that India now consumes the most antibiotics in the world. In 2010, India is estimated to have consumed 12.9 billion antibiotic pills. In comparison, China consumed 10 billion pills and the US, 6.8 billion. While antibiotic usage worldwide in the first decade of the 21st century rose by 36 per cent, in India, the count went up by 62 per cent, from 8 billion pills in 2001 to 12.9 billion in 2010. Seventy-six percent of the overall increase in global antibiotic consumption between 2000 and 2010 was attributable to BRICS countries, i.e., Brazil, Russia, India, China, and South Africa.
_
The scale-up in antibiotic use in India has been enabled by rapid economic growth and rising incomes, which have not translated into improvements in water, sanitation, and public health, although evidence exploring this key issue is anecdotal. Antibiotics continue to be prescribed or sold for diarrheal diseases and upper respiratory infections for which they have limited value. India’s large population is often blamed for the easy spread of resistant pathogens, but population densities in India are lower than those in parts of Indonesia or China. The main problem is that India lags on basic public health measures. Immunization rates (as measured by diphtheria-tetanus-pertussis [DPT3]) coverage in India (72%) lags behind those in Brazil (95%), China (99%), and Indonesia (85%). The percentage of the population with access to improved sanitation facilities in India (36%) was far lower than the percentage in Brazil (81.3%), China (65.3%), and Indonesia (58.8%). Under the Swacch Bharat Abhiyan (Clean India Program), the government has committed to providing toilets and improving sewage systems, but these measures will take time to implement.
_
The figure above shows that maximum isolates of E. Coli resistant to fluoroquinolone found in India:
_
New Delhi metallo-β-lactamase (NDM) enzymes, first reported in 2008, are now found worldwide. In India, Escherichia coli (n = 1,815) isolated from the community showed high overall resistance to ampicillin, naladixic acid, and co-trimoxazole (75%, 73%, and 59%, respectively) between 2004 and 2007. Nearly a third of isolates are resistant to injectables like aminoglycosides (represented by gentamicin). From 2008 to 2013, E. coli resistance to third-generation cephalosporins increased from 70% to 83%, and fluoroquinolone resistance increased from 78% to 85%. Ten percent of E. coli isolates were resistant to carbapenems in 2008, increasing to 13% in 2013. Among Klebsiella pneumoniae isolates, third-generation cephalosporin resistance decreased from 90% to 80%, and fluoroquinolone resistance increased from 57% to 73%. Carbapenem resistance among K. pneumoniae increased from 2% in 2002 to 52% in 2009 in one tertiary-care hospital in New Delhi. Resistance to fluoroquinolones among invasive Salmonella Typhi isolates in India increased from 8% in 2008 to 28% in 2014. However, resistance in 2014 to two older antibiotics—ampicillin, 5%, and cotrimoxazole, 4%—is decreasing, possibly because of a decline in consumption of these two drugs, and is much lower than rates of resistance to fluoroquinolones. Resistance to nalidixic acid in S. Typhi is increasing (resistance is about 20%–30%) because of widespread use of other quinolones and not because of nalidixic acid use per se. Among Enterococcus faecium isolates, 11% were vancomycin resistant. Surgical site infections are a problem and are predominantly related to Gram-negative pathogens. A recent study from Mumbai reported a 1.6% rate of surgical site infections, with 66% caused by Gram-negative bacilli (GNB). With diminishing options for treating multidrug-resistant Acinetobacter baumannii and other resistant infections, colistin use is increasing, but resistance to colistin is on the rise. Gram-positive infections are also a problem. High rates of methicillin-resistant Staphylococcus aureus (MRSA) in clinical isolates from various studies in India have been documented, with rates as high as 54.8% (ranging between 32% and 80%) recorded. A recent report records a steep increase in MRSA, from 29% of S. aureus isolates in 2009 to 47% in 2014 based on data from a large private laboratory network. A few weeks ago, a public health research organisation with offices in Washington and New Delhi, the Center for Disease Dynamics, Economics & Policy (CDDEP), published an in-depth report detailing antibiotic use and resistance across the world. It too had found India a hotbed for antibiotic resistance. The resistance of Klebsiella pneumonia to carbapenems, the antibiotic of last resort, for instance, was 57 per cent compared to just below 5 per cent across Europe. Resistance to other bacteria like E Coli, MRSA (Methicillin-resistant Staphylococcus aureus), and ESBL- producing bacteria (Extended-spectrum beta-lactamases) was just as bad as in other countries. The most vulnerable, of course, are newborns. According to a Lancet study last year, 58,000 infants in India are estimated to have died from bacterial infections in 2013.
_
Antibiotic resistance is a global phenomenon. But its epicentre is India. The country is afflicted by easy access to the strongest of antibiotics without prescriptions or diagnoses; by qualified doctors, not just quacks, who prescribe drugs with little thought; by hospitals where overuse has created colonies of these superbugs; by excessive usage on livestock; and by poor sanitation. All this has created a kind of perfect storm for these super-resistant microbes to menace our health. A large pharmaceutical industry, high background rates of infectious diseases and an affluent population that can afford antibiotics. You put all the things together and you get high pathogenic strains. As far as AMR is concerned, every individual is at the mercy of others, rich or poor. Antibiotics are different from other drugs. If someone was to take a statin, for instance, it’s not going to diminish the statin’s effectiveness for you. But with antibiotics, you can get an infection that is drug-resistant even if you’ve never misused antibiotics in your life.
__
Most of the Government hospitals in India are not worried about resistance. They are still struggling to get hold of life saving antibiotics. The private practitioners, private and corporate hospitals are the breeding grounds for resistance. There are very few hospitals in India with infectious diseases and infection control specialists. Only in a minority of Indian private hospitals are antibiotic policy and antibiotic stewardship implemented. The majority of private and corporate hospitals are in denial, either purposefully or due to ignorance. There are many hospital administrators in India claiming zero infection in their hospitals. It is sad to say that many of these hospitals do not have the necessary microbiology laboratory support or trained infection control specialists to look for resistance. The claim of zero infection is in fact an innocent advertisement of the lack of necessary infection control infrastructure in that hospital.
_
India awoke late to risks of antibiotic overuse and is scrambling to contain the surge in drug resistance. After years of doing little to tackle the silent but potentially deadly proliferation of antibiotic-resistant bacteria in India, all hell broke loose in 2008, when New Delhi was tacked onto the name of a one such bug. The New Delhi Metallo-beta-lactamase-1 was an enzyme that rendered bacteria resistant to a broad spectrum of antibiotics. A strain of the NDM1 had crossed the shores and spread resistance in the U.K. as well. Despite its outrage over being associated with a resistant bug, the nation sat up to the danger of anti-microbial resistance within its boundaries, and is beginning to understand the disastrous societal consequences of rendering certain life-saving drugs impotent. Prior to the detection of the NDM, isolated calls for regulating antibiotics use were being made by doctors in surgeries and those manning Intensive Care Units. It was in 2011 that Indian Union government came up with a National Policy for Containment of Antimicrobial Resistance in India, seeking to reverse what seemed to be spiralling healthcare concern. The policy makes no bones about recognising the real threat: Antimicrobial resistance in pathogens causing important communicable diseases has become a matter of great public health concern globally including our country. Resistance has emerged even to newer, more potent antimicrobial agents like carbapenems. Among the key factors responsible are the widespread use and availability of practically all the antimicrobials across the counter, increasing and wanton use of antibiotics in livestock production, inappropriate doses, and irrational use of antibiotics in hospitals. Attempts have begun to regulate at least the human consumption of antibiotics: there are now guidelines for appropriate antibiotics usage which have revised Schedule H drugs to make over-the-counter availability of certain antibiotics nearly impossible. Stringent enforcement of drugs control, making the dispensing of some antibiotics over the counter punishable, is the need of the hour.
___
India faces a twin challenge of overconsumption of antibiotics breeding drug-resistant bacteria while ensuring that the poor and vulnerable have easy access. So like many other developing countries, India has to turn the spotlight on ensuring sustainable access even while maintaining sustainable effectiveness of all antibiotics. The only way to achieve this twin objective is by ensuring that all stakeholders — government, patients, veterinarians, doctors, pharmacists, pharmaceutical companies and health-care facilities — play their respective roles more responsibly. First, people should be made aware that stopping antibiotics midway, missing doses, taking suboptimal dosages, or consuming antibiotics for cold and other viral infections, to name a few, makes them resistant to antibiotics; when ill the next time, their only recourse will be more expensive drugs or probably nothing at all. This is best exemplified in the case of multidrug-resistant tuberculosis that requires longer period of treatment using very toxic drugs that are more expensive. A much-needed public awareness campaign to highlight the dangers of misuse and irrational use of antibiotics was recently launched by the Ministry of Health and Family Welfare in 2016. Called ‘Medicines with the Red Line’, it comes at a time when the consumption of antibiotics in India has increased sharply while the effectiveness of these drugs to treat bacterial infections has been steadily declining. High disease burden, rising income, cheap, unregulated sales of antibiotics and poor public health infrastructure are some of the reasons for the sharp increase in antibiotic use. As a 2013 study in Indian Journal of Medical Ethics revealed, knowledge of antibiotic resistance was “reasonable among doctors, but low in priority”. Inadequate diagnostic facilities, lack of antibiotic guidelines and patients’ demand for quick relief often determined doctors’ prescription habits, besides incentives from drugs companies and chemists to push certain products. The collusion of drug companies and chemists is also apparent in the rampant over-the-counter (OTC) sale of antibiotics, particularly carbapenems (that is among the highest in the world), even for ailments where they are not indicative. The introduction of Schedule H1 category from March 2014 to prevent the sale of 24 third- and fourth-generation antibiotics without prescription is a step in the right direction. Licences of 213 retail pharmacies have been cancelled for non-compliance. But restricting OTC sales of antibiotics, particularly the commonly used ones, is a double-edged sword. Any intervention to limit access by enforcing prescription-only laws unwittingly cuts off a vast majority of the population, particularly in the rural areas, that lacks access to doctors.
_
Antibiotic use, environment and antibiotic resistance: A qualitative study among human and veterinary health care professionals in Orissa, India: a 2008 study:
The main finding of the study was mishandling and abuse of antibiotics in patients as well as at professional level due to weak implementation of legislation, which appears to be the major cause of antibacterial agent resistance. Incomplete course or dose due to poverty in rural area and self-medication in urban area are more common. The study also showed that climatic factors, pollution and population density are the major ecological factors which influence antibiotic prescriptions. Another major finding of this study was that, due to improper disposal system of pharmaceuticals; antibiotics are contaminating air, water and terrains which can cause major risk to aquatic and grazing animals.
_
Factors influencing primary care physicians to prescribe antibiotics in Delhi India: a 2010 study:
This is one of the first studies from developing countries to conduct an in-depth exploration of primary care doctors’ views on their antibiotic prescribing, resistance and interventions for decrease in antibiotic misuse and resistance. Findings from this study revealed that primary care doctors prescribe antibiotic mainly in the following contexts: (i) diagnostic uncertainty, (ii) perceived demand or expectations from patient, (iii) to retain patients and gain financially, (iv) under the influence of medical representatives and (v) under the misguided impression that the poorer section of society needs antibiotics due to unhygienic living conditions. Factors that led public sector doctors to prescribe antibiotics were (i) diagnostic uncertainty due to the absence of a lab facility, (ii) patient expectations to get ‘capsules’, (iii) lack of time to interact with patients and (iv) oversupplied and near-expiry antibiotics. Doctors also stressed that due to laxity in rules and regulations for antibiotic prescribing and dispensing, chemists, patients and ‘quacks’ misuse antibiotics. Many doctors felt that prescribing the latest antibiotics not only removed their insecurity but the patients also were satisfied that their visit had been useful as they were being prescribed expensive and new antibiotics, with an obvious expectation of better efficacy. This is what Kunin et al. have called ‘drugs of fear’ that characterize the compelling need of physicians to use the latest and best antibiotics to solve a problem and to meet patient’s expectations. A qualitative study conducted in UK has shown that many times GPs prescribe antibiotics to their patients as they believe it is their duty to do the best for them and are concerned about more serious problems rather than the theoretical complications of antimicrobial resistance. Family Physicians in the USA have also argued their antibiotic prescribing decision as balancing act. These findings are in agreement with author’s findings and suggest that prescribers in different settings respond and behave in a comparable way to complex challenges. Doctors also pointed out that once they have written a prescription for a particular symptom, the same prescription is used for self-medication to save money and time required for repeated consultations. This behaviour of repeating the prescription, purchasing antibiotics without a prescription and purchasing fewer units than prescribed is also reported by Lansang et al. from Manila. On the contrary, dispensing a full course of antibiotics and fixed-size packs in the developed world has also shown that patients keep the unused antibiotics and use the leftover antibiotics at a later time. Another factor that influences which antibiotic is prescribed and overuse of antibiotics in the private sector was found to be medical representatives’ pressure. Influence of pharmaceutical companies on doctors is very well recognized and proceedings of legal cases provide insight into the extent of influence this industry can impose on doctors and other stakeholders. This has been revealed by Søndergaard et al. in their study, which shows that pharmaceutical representative’s visits markedly influence doctor’s drug preference towards the marketed drug.
_
Antibiotic use and resistance: perceptions and ethical challenges among doctors, pharmacists and the public in Vellore, South India: a 2013 study:
It was found that the public had minimal awareness of resistance, antibiotics and infections. They wanted symptomatic relief. Doctors reported prescribing antibiotics for perceived patient expectations and quick recovery. Business concerns contributed to antibiotics sales among pharmacists. Pharmaceutical industry incentives and healthcare provider competition were the main ethical challenges. Suggested interventional strategies by the participants included creating public awareness, better healthcare provider communication, improved diagnostic support, strict implementation of guidelines, continuing education, and strengthening of regulations.
__
Global study reveals soaring antibiotic resistance in India: a 2015 study:
The first in-depth look at antimicrobial resistance worldwide has uncovered the extent to which antibiotics are misused, particularly in increasingly prosperous countries, including India, Vietnam and Kenya. The investigators say preventing resistance to existing antibiotics should take priority over developing new drugs, since they will also fail if the causes of resistance are not tackled. The new analysis includes data on drug resistance in 39 countries, and profiles of antibiotic use in 69 countries, and is the first time such data has been combined. Previous reports have focused only on resistance, and included only data from public sources. “Much of this data has never seen the light of day before because we dug it out from private clinics in these middle-income countries like India,” says Ramanan Laxminarayan of the Center for Disease Dynamics, Economics and Policy in New Delhi, India. “It’s the first global snapshot of antibiotic use and resistance.” The analysis reveals soaring rates of resistance in countries of growing wealth, especially India, where more people are demanding antibiotics for minor infections, and resistance rates among bacteria are soaring. “We’ve seen a huge increase in MRSA in India, from 29 per cent of isolates in 2009 to 47 per cent in 2014,” says Laxminarayan. Equally alarming, he says, is a surge in Klebsiella pneumoniae, which can cause fatal lung infections. It is resistant to Carbapenems, an antibiotic that is used as a last resort. In 2014, 57 per cent of samples tested in India were resistant, compared with virtually none six years ago. “These bugs weren’t a problem at all, but now we stand on the brink of almost losing a whole class of vital antibiotics,” says Laxminarayan.
___
Antibiotic Resistance in India: Drivers and Opportunities for Action: a 2016 Indian study:
Summary Points:
1. Antibiotic use is a major driver of resistance. In 2010, India was the world’s largest consumer of antibiotics for human health.
2. Access to antibiotics is rising, which portends well for the large proportion of India’s population that has thus far had poor access to these life-saving drugs.
3. The convergence of factors such as poor public health infrastructure, rising incomes, a high burden of disease, and cheap, unregulated sales of antibiotics has created ideal conditions for a rapid rise in resistant infections in India.
4. Over-the-counter, nonprescription sales of carbapenems in India are among the highest in the world and contribute to growing carbapenem resistance among Gram-negative organisms.
5. Improving regulation of drug production and sales, better managing physician compensation, and encouraging behavior change among doctors and patients are of immediate priority.
____
How dirty production of drugs helps create superbugs in India:
Scientists have been saying for years that polluting pharma plants are directly contributing to the spread of antibiotic resistance. Now, it looks as if they could have hard evidence in India, the world’s biggest antibiotic producer. A hard-hitting report by campaigning organization Changing Markets says direct sampling of water from manufacturing sites operated by Aurobindo, Orchid Pharma and Asiatic Drugs and Pharmaceuticals has–for the first time–uncovered drug-resistant bacteria. The researchers behind the work say the testing proves pharma pollution ranks alongside excessive consumption of antibiotics in human medicine and profligate use in livestock as a key driver for resistance. And their focus on Indian producers is warranted, as India is the world’s largest maker–and consumer–of antibiotics. Worryingly, out of 34 Indian manufacturing sites tested, almost half (16) were harboring resistant strains of bacteria. At four of these, resistance was detected to three widely-used and important antibiotic classes–cephalosporins, carbapenems and fluoroquinolones–including drugs generally reserved for use as a last resort. At eight sites, resistance to cephalosporins and fluoroquinolones was detected, while another four sites had evidence of either cephalosporin or fluoroquinolone resistance. This suggests industrial waste containing active antibiotic ingredients is being leaked into the surrounding environment. The release of untreated waste products from substandard antibiotic factories “acts as a driver for the development of drug resistance, creating environmental ‘reservoirs’ of antibiotic-resistant bacteria,” said the report. Studies have shown how this causes nearby bacteria to develop immunity to the drugs – creating “superbugs” – and that those resistant bacteria then spread around the world. Various studies have found “high levels of hazardous waste” and “large volumes of effluent waste” being dumped into the environment surrounding factories in India and China, where most of the world’s antibiotics are produced. Active ingredients used in antibiotics get into the local soil and water systems, leading to bacteria in the environment becoming resistant to the drugs. Once established in the environment, the bacteria can exchange genetic material with nearby germs. They can then spread around the world through air and water, and by travellers visiting countries where the bacteria are prevalent.
____
India has indeed become epicentre of AMR due to following reasons:
a) Antibiotic use is a major driver of resistance. India is the world’s largest consumer of antibiotics for human health.
b) Over-the-counter, non-prescription sale of carbapenems is highest in the world in India and contribute to growing carbapenem resistance among Gram-negative organisms. India is afflicted by easy access to the strongest of antibiotics without prescriptions or diagnoses; by qualified doctors, not just quacks, who prescribe drugs with little thought; and by hospitals where overuse, misuse and inappropriate doses have created colonies of superbugs.
c) Release of untreated waste products containing active antibiotic ingredients from many substandard Indian pharmaceutical factories contaminate air, water and terrains that acts as a driver for the development of drug resistance and create environmental ‘reservoirs’ of antibiotic-resistant bacteria. Once established in the environment, the resistant bacteria can exchange genetic material with nearby bacteria and then spread around the world through air and water, and by travellers.
d) Growing wealth where many people are demanding antibiotics for minor infections coupled with cheap antibiotics result in inappropriate use of antibiotics.
e) India lags behind on basic public health measures like immunization, clean drinking water, vector control and sanitation; coupled with overcrowding, malnutrition, poor personal hygiene and an excess of ‘alternative medicine’ options result in high rates of infectious diseases which necessitates greater antibiotic use. Also, antibiotics continue to be prescribed or sold for diarrheal diseases and upper respiratory infections for which they have limited value.
f) Incomplete course or dose due to poverty in rural area and self-medication in urban area are common.
g) Excessive and unregulated usage of antibiotics in livestock and animal food production contributes to resistance in microorganisms.
h) Most Indian hospitals have no antibiotic policy, no antibiotic stewardship and no infection control specialists.
____
____
Think of world without antibiotics due to AMR:
•Without antibiotics, there would be very little elective surgery. Before sulfa drugs, surgery was a very serious business with a high risk that a patient might die of some complicating infection.
•Without antibiotics, forget organ transplants. The immune suppression would almost certainly be fatal in a pretty short time period. HIV would also be more dangerous.
•Without antibiotics, retirements would get shorter again. Before antibiotics, the average 60 year old who caught pneumonia was more likely than not to die of it.
•Without antibiotics, maternal mortality would be a lot higher and mortality from abortions increased.
•The severely disabled would have much shorter life spans. Without antibiotics, there would be no way to treat the bed sores, or the lung and urinary tract infections that are common for people with limited sensation or mobility.
•Strep and its evil cousins, scarlet and rheumatic fevers, would once again be a major killer and disabler of children.
_____
_____
Mechanisms of Action of Antimicrobial Agents:
_
The figure below shows mechanisms of action of antibiotics on various bacteria:
_
In order to appreciate the mechanisms of resistance, it is important to understand how antimicrobial agents act. Antimicrobial agents act selectively on vital microbial functions with minimal effects or without affecting host functions. Different antimicrobial agents act in different ways. The understanding of these mechanisms as well as the chemical nature of the antimicrobial agents is crucial in the understanding of the ways how resistance against them develops. Broadly, antimicrobial agents may be described as either bacteriostatic or bactericidal. Bacteriostatic antimicrobial agents only inhibit the growth or multiplication of the bacteria giving the immune system of the host time to clear them from the system. Complete elimination of the bacteria in this case therefore is dependent on the competence of the immune system. Bactericidal agents kill the bacteria and therefore with or without a competent immune system of the host, the bacteria will be dead. However, the mechanism of action of antimicrobial agents can be categorized further based on the structure of the bacteria or the function that is affected by the agents. Three conditions must be met for an antibiotic to be effective against bacteria: i) a susceptible antibiotic target must exist in the cell, ii) the antibiotic must reach the target in sufficient quantity, and iii) the antibiotic must not be inactivated or modified. Understanding antibiotic resistance mechanisms requires an understanding of where antibiotics exert their effect. There are five major modes of antibiotic mechanisms of activity and here are some examples.
1. Interference with cell wall synthesis:
B-lactam antibiotics such as penicillins and cephalosporins interfere with enzymes required for the synthesis of the peptidoglycan layer. Glycopeptides (vancomycin, teicoplanin, oritavancin) target the bacterial cell wall by binding to the D-alanyl-D-alanine termini of the peptidoglycan chain, thereby preventing the cross-linking steps. Telavancin, a novel rapidly bactericidal lipoglycopeptide, inhibits peptidoglycan biosynthesis through preferential targeting of transglycosylation.
2. Inhibition of protein synthesis:
Macrolides bind to the 50S ribosomal subunit and interfere with the elongation of nascent polypeptide chains. Aminoglycosides inhibit initiation of protein synthesis and bind to the 30S ribosomal subunit. Chloramphenicol binds to the 50S ribosomal subunit blocking peptidyltransferase reaction. Tetracyclines inhibit protein synthesis by binding to 30S subunit of ribosome, thereby weakening the ribosome-tRNA interaction. The semisynthetic tetracycline derivatives, colloquially termed the glycylglycines, act at the bacterial ribosome to arrest translation. The glycylglycines bind the ribosome more tightly than previous tetracyclines, so that the TetM resistance factor is unable to displace them from this site, hence TetM is unable to protect the ribosomes from the action of these new drugs. The TetA-mediated efflux system is ineffective against the glycylglycines, as they are not substrates for the transporter. The oxazolidinones, one of the newest classes of antibiotics, interact with the A site of the bacterial ribosome where they should interfere with the placement of the aminoacyl-tRNA.
3. Interference with nucleic acid synthesis:
Rifampicin interferes with a DNA-directed RNA polymerase. Quinolones disrupt DNA synthesis by interference with type II topoisomerases DNA gyrase and topoisomerase IV during replication and by causing double strand breaks.
4. Inhibition of a metabolic pathway:
The sulfonamides (e.g. sulfamethoxazole) and trimethoprim each block the key steps in folate synthesis, which is a cofactor in the biosynthesis of nucleotides, the building blocks of DNA and RNA.
5. Disorganizing of the cell membrane:
The primary site of action is the cytoplasmic membrane of Gram-positive bacteria, or the inner membrane of Gram-negative bacteria. It is postulated that polymyxins exert their inhibitory effects by increasing bacterial membrane permeability, causing leakage of bacterial content. The cyclic lipopeptide daptomycin displays rapid bactericidal activity by binding to the cytoplasmic membrane in a calcium-dependent manner and oligomerizing in the membrane, leading to an efflux of potassium from the bacterial cell and cell death.
_
A summary of the mode of action for the major classes of antibiotics is provided in table below:
______
______
Mechanisms of antimicrobial resistance (AMR):
In simplifying the resistance phenomenon, we can focus on two factors: the antibiotic, which, acting as a selective agent, helps to propagate organisms that have the second factor: the resistance gene. If either the antibiotic or the resistance gene were not present, we would not face a resistance problem. The finding of resistance to a new agent in an organism is not unexpected, since antibiotics and other related organic molecules are, or resemble, natural products. Over their many millennia of existence, bacteria have continuously confronted organic structures that affect their growth; to survive, bacteria have acquired resistance genes. It is, however, the appearance of these resistance genes in a clinical isolate and in a clinical setting that is a warning to clinicians to control use of the new drug. In this regard, the discovery of resistance in a bacterial strain, for instance a commensal, portends future problems with resistance in clinical pathogens in that hospital or community. Attention should be focused on the use of the drug and the spread of resistance genes. The abilities of bacterial organisms to utilize the various strategies to resist antimicrobial compounds are all genetically encoded. Bacteria may be intrinsically resistant to ≥1 class of antimicrobial agents, or may acquire resistance by de novo mutation or via the acquisition of resistance genes from other organisms. Acquired resistance genes may enable a bacterium to produce enzymes that destroy the antibacterial drug, to express efflux systems that prevent the drug from reaching its intracellular target, to modify the drug’s target site, or to produce an alternative metabolic pathway that bypasses the action of the drug. Acquisition of new genetic material by antimicrobial-susceptible bacteria from resistant strains of bacteria may occur through conjugation, transformation, or transduction, with transposons often facilitating the incorporation of the multiple resistance genes into the host’s genome or plasmids. Use of antibacterial agents creates selective pressure for the emergence of resistant strains. Non-genetic reasons why bacteria may not be inhibited by antibiotics are that drugs may not reach bacteria located in the center of an abscess and that certain drugs, such as penicillin, will not affect bacteria that are not growing (intrinsic resistance). Also the presence of foreign bodies makes successful antibiotic treatment more difficult.
____
Selection pressure:
_
Antimicrobial resistance can be caused by “selection pressure.” The increased prevalence and dissemination of resistance is an outcome of natural selection and should be viewed as an expected phenomenon of the Darwinian biological principle of “survival of the fittest.” In any large population of bacteria, a few cells will be present which possess traits that enable them to survive in the presence of a noxious substance, in this case the ability to fend off the action of the antimicrobial. Regardless of how effective an antimicrobial might be, rarely, if ever, will 100% of the organisms be killed during a course of treatment. This means that at least one organism out of thousands may have developed resistance to the antimicrobial. The few surviving and potentially resistant organisms could then transfer their genetic material to offspring or even other unrelated organisms as a striking example of the evolutionary principle of natural selection. Bacteria are extremely numerous and remarkably prolific. If conditions are right, a single bacterium can divide rapidly and produce a billion offspring in a single day. Surviving microbes become dominant, adapting quickly to changing environmental conditions, including the introduction of new antimicrobial agents. Furthermore, multiple resistance mechanisms in a single bacterial pathogen are becoming the norm. This complicates treatment of these infections, increasing both morbidity and healthcare costs.
_
Resistance among bacteria poses a distinct threat:
Resistance is, of course, the evolutionary consequence of the deployment of a selective pressure; therefore, it has been documented among all organisms against which we have declared biological wars, from viruses to insects. However, resistance among bacteria poses a distinct threat because of various reasons: (a) the abuse of antibacterial drugs is much higher than that of antifungal or antiviral agents; the later ones are seldom self-prescribed, wrongfully used as prophylaxis, or have agricultural usage; (b) bacterial genetic characteristics and abilities enable a rapid evolution toward resistance in ways that exceed by far those of viruses, fungi, and protozoa: haploidy, horizontal gene transfer mechanisms, extrachromosomal elements are all features that foster resistance and that are almost unique to bacteria; (c) bacteria appear to be much more abundant than viruses, fungi, and protozoa as microbiota of humans, which increases exponentially the exposure of the former to antibiotics each time they are used clinically, creating more chances of resistance to emerge and be selected; (d) bacterial diseases are also more abundant, at least for treatment purposes, increasing also the exposure to antibacterial drugs, perhaps with the exception of malaria. Therefore, although microbial resistance in general is posing grave problems for public health, it is not possible to view all resistance from the same perspective.
_
_
Antibiotic resistance is not a new phenomenon. Although reported only since the 1940s, resistance has existed in nature for thousands of years. It has evolved as bacteria, fungi and parasites spontaneously produce antibacterial, antiparasitic and antifungal substances in order to survive in competition with these other species. In fact, this is how many of the antibiotics used today — for instance penicillin — were discovered. However, over the last 50 years, an increasing number of bacterial species have developed resistance to antibiotics. Moreover, genetic analysis has revealed that certain resistance genes have recently emerged as a consequence of overuse and misuse of antibiotics in medical and veterinary settings. As the global consumption of antibiotics as well as international travel continue to rise, bacteria are mutating and exchanging their resistance genes at an unprecedented pace. Bacteria have the natural ability to multiply and change their genetic material (which we call “mutate”) very quickly, which can be seen as a survival mechanism that allows them to adapt to new environments. Every time we take antibiotics — or use them in animals — we create a selection pressure on resistant bacteria to survive and give them an opportunity to adapt to antibiotics.
_
Biology of Antibiotic Resistance:
Understanding the mechanisms of resistance has become a significant biochemical issue over the past several years and nowadays there is a large pool of information about how bacteria can develop drug resistance. Biochemical and genetic aspects of antibiotic resistance mechanisms in bacteria are shown in the figure below:
The figure above shows genetic and biochemical aspects of acquired antibiotic resistance mechanisms in bacteria.
__
Genetics of Antibiotic Resistance:
Antimicrobial resistance traits are genetically coded, and can either be intrinsic or acquired.
•Intrinsic resistance is due to innately coded genes which create natural “insensitivity” to a particular antibiotic. Innate resistance is normally expressed by virtually all strains of that particular bacterial species. While expressing intrinsic resistance, microorganisms naturally do not possess target sites for the drugs and therefore the drug does not affect them or they naturally have low permeability to those agents.
•Acquired resistance is gained by previously susceptible bacteria either through mutation or horizontally obtained from other bacteria possessing such resistance via transformation, transduction or conjugation. Acquired resistance is limited to subpopulations of a particular bacterial species and may result from selective pressure exerted by antibiotic usage.
_
Studies of a wide variety of bacterial pathogens have identified numerous genetic loci associated with antibiotic resistance. For some types of resistance there is a large diversity of responsible genetic determinants. Resistance can be an intrinsic property of the bacteria themselves or it can be acquired. Acquired bacterial antibiotic resistance can result from a mutation of cellular genes, the acquisition of foreign resistance genes or a combination of these two mechanisms. Thus, there are two main ways of acquiring antibiotic resistance: i) through mutation in different chromosomal loci and ii) through horizontal gene transfer (i.e. acquisition of resistance genes from other microorganisms). This raises several questions about the evolution and ecology of antibiotic resistance genes. Phylogenetic insights into the evolution and diversity of several antibiotic resistance genes suggest that at least some of these genes have a long evolutionary history of diversification that began well before the ‘antibiotic era’.
__
Mutation:
A mutation is a spontaneous change in the DNA sequence within the gene that may lead to a change in the trait which it codes for. Any change in a single base pair may lead to a corresponding change in one or more of the amino acids for which it codes, which can then change the enzyme or cell structure that consequently changes the affinity or effective activity of the targeted antimicrobials. In prokaryotic genomes, mutations frequently occur due to base changes caused by exogenous agents, DNA polymerase errors, deletions, insertions and duplications. For prokaryotes, there is a constant rate of spontaneous mutation of about 0.0033 mutations per genome per cell generation that is relatively uniform for a diverse spectrum of organisms. The mutation rate for individual genes varies significantly among and within genes.
Random mutations:
Mutations are random. Mutations can be beneficial, neutral, or harmful for the organism, but mutations do not “try” to supply what the organism “needs.” Factors in the environment may influence the rate of mutation but are not generally thought to influence the direction of mutation. For example, exposure to harmful chemicals may increase the mutation rate, but will not cause more mutations that make the organism resistant to those chemicals. In this respect, mutations are random — whether a particular mutation happens or not is unrelated to how useful that mutation would be. For example, in the U.S. where people have access to shampoos with chemicals that kill lice, we have a lot of lice that are resistant to those chemicals. There are two possible explanations for this:
Scientists generally think that the first explanation is the right one and that directed mutations, the second possible explanation relying on non-random mutation, is not correct. Researchers have performed many experiments in this area. Though results can be interpreted in several ways, none unambiguously support directed mutation. Nevertheless, scientists are still doing research that provides evidence relevant to this issue. In addition, experiments have made it clear that many mutations are in fact random, and did not occur because the organism was placed in a situation where the mutation would be useful. For example, if you expose bacteria to an antibiotic, you will likely observe an increased prevalence of antibiotic resistance. Esther and Joshua Lederberg determined that many of these mutations for antibiotic resistance existed in the population even before the population was exposed to the antibiotic — and that exposure to the antibiotic did not cause those new resistant mutants to appear.
The Lederberg experiment:
In 1952, Esther and Joshua Lederberg performed an experiment that helped show that many mutations are random, not directed. In this experiment, they capitalized on the ease with which bacteria can be grown and maintained. Bacteria grow into isolated colonies on plates. These colonies can be reproduced from an original plate to new plates by “stamping” the original plate with a cloth and then stamping empty plates with the same cloth. Bacteria from each colony are picked up on the cloth and then deposited on the new plates by the cloth. Esther and Joshua hypothesized that antibiotic resistant strains of bacteria surviving an application of antibiotics had the resistance before their exposure to the antibiotics, not as a result of the exposure. So the penicillin-resistant bacteria were there in the population before they encountered penicillin. They did not evolve resistance in response to exposure to the antibiotic.
_
Spontaneous mutation in the bacterium’s DNA:
Mutations—spontaneous changes in a bacterium’s genetic material—occur in about one in one million to one in ten million cells. Mutations are rare but the fact that bacteria reproduce at such a high rate allows for the effect to be significant. Even if only a single S. aureus cell were to make its way into your wound, it would take only 10 generations for that single cell to grow into a colony of more than 1,000 (210 = 1,024), and just 10 more generations for it to erupt into a colony of more than 1 million (220 = 1,048,576). For a bacterium that divides about every half hour (which is how quickly S. aureus can grow in optimal conditions), that is a lot of bacteria in less than 12 hours. S. aureus has about 2.8 million nucleotide base pairs in its genome. At a rate of, say, 10-10 mutations per nucleotide base, that amounts to nearly 300 mutations in that population of bacteria within 10 hours! To better understand the impact of this situation, think of it this way: With a genome size of 2.8 × 106 and a mutation rate of 1 mutation per 1010 base pairs, it would take a single bacterium 30 hours to grow into a population in which every single base pair in the genome will have mutated not once, but 30 times! Thus, any individual mutation that could theoretically occur in the bacteria will have occurred somewhere in that population—in just over a day. Depending on the mutation, different types of resistance can occur.
For example, mutations can:
1. provide the ability to close or seal up the entry ports through which antibiotics enter a bacterial cell
2. modify the bacterial cell target that the antibiotic is programmed to attack
3. increase the production of natural enzymes that inactivate antibiotics
4. allow the bacteria to increase the production of natural pumping mechanisms that expel the antibiotic for the bacterial cell so that it fails to reach its target.
These mutations enable the bacteria to survive or resist the effects of the antibiotic. These ‘new and improved’ model bacteria stay alive and continue to replicate and multiply. Remember these spontaneous mutations are random and did not arise in response to exposure to the antibiotic. Those bacteria with a mutation that allows them to survive live to reproduce. They then pass this trait to their offspring, which leads to the evolution of a fully resistant colony. Although these chromosomal mutations may seem to benefit the bacteria by providing antibiotic resistance, they also confer a cost of fitness. For example, a ribosomal mutation may protect a bacterial cell by changing the binding site of an antibiotic but it will also slow the process of protein synthesis. Additionally, a particular study specifically compared the overall fitness of antibiotic resistant strains of Escherichia coli and Salmonella typhimurium to their drug-sensitive revertants. They observed a reduced overall fitness in the antibiotic resistant strains, especially in growth rate. Antibacterial resistance may impose a biological cost, thereby reducing fitness of resistant strains, which can limit the spread of antibacterial-resistant bacteria, for example, in the absence of antibacterial compounds. Additional mutations, however, may compensate for this fitness cost and can aid the survival of these bacteria. Paleontological data show that both antibiotics and antibiotic resistance are ancient compounds and mechanisms. Useful antibiotic targets are those for which mutations negatively impact bacterial reproduction or viability.
_
When a susceptible bacterium comes into contact with a therapeutic concentration of antimicrobials, like fluroquinolones, the antimicrobial can bind to the specific enzymes, in this case, DNA gyrase. The DNA gyrase is an essential bacterial enzyme required for DNA replication. The end result is that fluoroquinolones block bacterial DNA replication leading to cell death. However, when spontaneous mutations occur in specific areas of the genes encoding these enzymes, antimicrobials no longer bind efficiently. This allows the bacterium to continue DNA replication.
_
For a long time, it has been thought that, for a microorganism to become resistant to an antibiotic, it must be in a large population. However, recent findings show that there is no necessity of large populations of bacteria for the appearance of antibiotic resistance. We know now that small populations of E.coli in an antibiotic gradient can become resistant. Any heterogeneous environment with respect to nutrient and antibiotic gradients may facilitate the development of antibiotic resistance in small bacterial populations. Researchers hypothesize that the mechanism of resistance development is based on four SNP (single-nucleotide polymorphism) mutations in the genome of E.coli produced by the gradient of antibiotic. These mutations confer the bacteria emergence of antibiotic resistance.
__
The development of resistance is inevitable following the introduction of a new antibiotic. Initial rates of resistance to new drugs are normally on the order of 1%. However, modern uses of antibiotics have caused a huge increase in the number of resistant bacteria. In fact, within 8-12 years after wide-spread use, strains resistant to multiple drugs become widespread. Multiple drug resistant strains of some bacteria have reached the proportion that virtually no antibiotics are available for treatment. Antibiotic resistance in bacteria may be an inherent trait of the organism (e.g. a particular type of cell wall structure) that renders it naturally resistant, or it may be acquired by means of mutation in its own DNA or acquisition of resistance-conferring DNA from another source.
______
Vertical vs. horizontal gene transfer:
_
Once the resistance genes have developed by mutation, they are transferred directly to all the bacteria’s progeny during DNA replication. This is known as vertical gene transfer or vertical evolution. The process is strictly a matter of Darwinian evolution driven by principles of natural selection: a spontaneous mutation in the bacterial chromosome imparts resistance to a member of the bacterial population. In the selective environment of the antibiotic, the wild types (non-mutants) are killed and the resistant mutant is allowed to grow and flourish. In the case of bacteria, resistant genes may be also acquired ‘horizontally’ across two bacteria of the same generation through the exchange of mobile genetic elements. Bacteria can develop resistance to antibiotics by mutating existing genes (vertical evolution), or by acquiring new genes from other strains or species (horizontal gene transfer). The sharing of genes between bacteria by horizontal gene transfer occurs by many different mechanisms. Mobile genetic elements, including phages, plasmids and transposons mediate this transfer, and in some circumstances the presence of low levels of the antibiotic in the environment is the key signal that promotes gene transfer, perhaps ensuring that the whole microbial community is protected from the antibiotic. In horizontal gene transfer, newly acquired DNA is incorporated into the genome of the recipient through either recombination or insertion. Recombination essentially is the regrouping of genes, such that native and foreign (new) DNA segments that are homologous are edited and combined. Insertion occurs when the foreign DNA introduced into a cell shares no homology with existing DNA. In this case, the new genetic material is embedded between existing genes in the recipient’s genome. Horizontal gene transfer must overcome several barriers before stable maintenance of the transferred gene in the recipient bacteria can occur. DNA entry, avoidance of restriction systems, and incorporation into the host replication machinery is necessary steps for transfer. Although fulfilling these criteria permits a gene to transfer, the gene must also confer a selective advantage to expand within the bacterial population.
_
Once bacteria acquires resistant gene by mutation, it is passed on to its progeny by vertical gene transfer. Once bacteria acquires resistant gene from horizontal gene transfer, it is passed on to other bacteria of same species and different species by horizontal gene transfer. However, can bacteria transfer plasmids acquired through horizontal gene transfer vertically to its progeny?
Yes.
Here are some studies showing vertical transmission of plasmids:
Study-1
Vertical Transmission of Biosynthetic Plasmids in Aphid Endosymbionts (Buchnera): 2001:
This study tested for horizontal transfer of plasmids among Buchnera aphidicola strains associated with ecologically and phylogenetically related aphid hosts (Uroleucon species). Phylogenetic congruence of Buchnera plasmid (trpEG and leuABC) and chromosomal (dnaN and trpB) genes supports strictly vertical long-term transmission of plasmids, which persist due to their contributions to host nutrition rather than capacity for infectious transfer. Synonymous divergences indicate elevated mutation on plasmids relative to chromosomal genes.
Study-2
First report on vertical transmission of a plasmid DNA in freshwater prawn, Macrobrachium rosenbergii: 2014:
This study demonstrates vertical transmission of a plasmid DNA in a decapod Macrobrachium rosenbergii for the first time. Females at three different maturation stages (immature, matured and berried) and mature males were injected with a plasmid DNA and allowed to spawn with untreated counterparts. Using specific primers the plasmid DNA could be amplified from the offspring of all groups except that of berried females. For this confirmation genomic DNA was isolated from 3 pools of 10 post larvae in each group. This presents an ideal strategy to protect young ones at zero stress.
Study-3
Vertical transmission of Escherichia coli carrying plasmid-mediated AmpC (pAmpC) through the broiler production pyramid: 2014:
In Sweden the prevalence of Enterobacteriaceae with transferable resistance to extended-spectrum cephalosporins (ESCs) is low. However, in broilers ESC-resistant Escherichia coli is common, with a clear dominance of blaCMY-2. Antimicrobials are rarely used in broiler production in Sweden and cephalosporins are never used. Introduction through imported breeding stock and subsequent vertical transmission of the bacteria through the production pyramid could be one explanation for this high prevalence. E. coli carrying blaCMY-2 is frequently present among grandparent animals imported to Sweden for breeding purposes. The occurrence of one clone in all levels of the production pyramid indicates that its introduction through imported breeding stock and vertical transmission through the production pyramid could be one explanation for the high proportion of Swedish broilers colonized with ESC-resistant E. coli.
Study-4
Vertical transmission of highly similar bla CTX-M-1-harboring IncI1 plasmids in Escherichia coli with different MLST types in the poultry production pyramid: 2014:
The purpose of this study was to characterize sets of extended-spectrum -lactamases (ESBL)-producing Enterobacteriaceae collected longitudinally from different flocks of broiler breeders, meconium of 1-day-old broilers from theses breeder flocks, as well as from these broiler flocks before slaughter. Five sets of ESBL-producing Escherichia coli were studied by multi-locus sequence typing (MLST), phylogenetic grouping, PCR-based replicon typing and resistance profiling. The bla CTX-M-1-harboring plasmids of one set (pHV295.1, pHV114.1, and pHV292.1) were fully sequenced and subjected to comparative analysis. Eleven different MLST sequence types (ST) were identified with ST1056 the predominant one, isolated in all five sets either on the broiler breeder or meconium level. Plasmid sequencing revealed that bla CTX-M-1 was carried by highly similar IncI1/ST3 plasmids that were 105 076 bp, 110 997 bp, and 117 269 bp in size, respectively. The fact that genetically similar IncI1/ST3 plasmids were found in ESBL-producing E. coli of different MLST types isolated at the different levels in the broiler production pyramid provides strong evidence for a vertical transmission of these plasmids from a common source (nucleus poultry flocks).
_
All these studies show that plasmid carrying new gene can be incorporated in host cell genome and fixed so that vertical gene transfer to progeny can be facilitated. I hypothesize that resistance gene acquired through mutation which is transferred to progeny by vertical gene transfer; may replicate a copy that may be incorporated in the bacterial plasmid to be transmitted to other bacteria by horizontal gene transfer.
___
Lateral or horizontal gene transfer (HGT) is a process whereby genetic material contained in small packets of DNA can be transferred between individual bacteria of the same species or even between different species. There are at least three possible mechanisms of HGT, equivalent to the three processes of genetic exchange in bacteria. These are transduction, transformation or conjugation.
1. Conjugation occurs when there is direct cell-cell contact between two bacteria (which need not be closely related) and transfer of small pieces of DNA called plasmids takes place. Plasmid is a small, circular DNA molecule that is separate from and can replicate independently of the bacterial genome. Plasmids are assisted by small, mobile DNA elements called transposons. This is thought to be the main mechanism of HGT.
2. Transformation is a process where parts of DNA are taken up by the bacteria from the external environment. This DNA is normally present in the external environment due to the death and lysis of another bacterium.
3. Transduction occurs when bacteria-specific viruses (bacteriophages) transfer DNA between two closely related bacteria.
_
The figure below shows mechanisms of horizontal gene transfer:
a) Transformation occurs when naked DNA is released on lysis of an organism and is taken up by another organism. The antibiotic-resistance gene can be integrated into the chromosome or plasmid of the recipient cell.
b) In transduction, antibiotic-resistance genes are transferred from one bacterium to another by means of bacteriophages and can be integrated into the chromosome of the recipient cell (lysogeny).
c) Conjugation occurs by direct contact between two bacteria: plasmids form a mating bridge across the bacteria and DNA is exchanged, which can result in acquisition of antibiotic-resistance genes by the recipient cell. Transposons are sequences of DNA that carry their own recombination enzymes that allow for transposition from one location to another; transposons can also carry antibiotic-resistance genes.
Not all organisms have all three mechanisms, but each one helps to amplify the resistance determinant within the microbial world. The microbial environment has carried these various gene distribution systems over evolutionary periods, using them to defend itself against threats to its existence, such as those posed by antibiotics.
_
Figure below shows MDR plasmid:
Plasmids are typically circular pieces of DNA that are considered mobile because they can be passed between bacteria via conjugation, a process that briefly connects the cytoplasm of two bacteria allowing horizontal gene transfer. Antimicrobial resistance genes encoded in bacterial chromosomes, such as multidrug efflux pumps in P aeruginosa, are usually not mobile, whereas those that are carried in plasmids can disseminate rapidly among bacteria of the same or different species. Moreover, plasmids often carry multiple AMR genes. At times, these additional genes are acquired through transposable elements, or transposons. Transposons are mobile DNA sequences that can integrate either into the bacterial chromosome or a plasmid, often carrying AMR genes.
_
Genetic Vectors of Resistance Genes:
Resistance genes are most often encoded in extrachromosomal genetic elements or in segments that appear to have been recombined into the chromosome from other genomes. The largest of the extrachromosomal elements are the plasmids, which are self-replicating, double-stranded circles of DNA, some of which express mechanisms that transfer the plasmid to another bacterial cell. Bacteria isolated from patients 80 years ago or more, before antimicrobials were first used, had plasmids similar to those seen now, but then, the plasmids had no resistance genes. Resistance genes encoded in plasmids are often located within segments called transposons. Functioning transposons include transposases that enable the transposon to recombine into other genomes; defective transposons have lost that capability. Such recombination can be demonstrated in vitro; evidence in vivo is provided by transposons with identical nucleotide sequences on a variety of different plasmids. Resistance genes are often further clustered within elements called integrons, which are frequently found within transposons and plasmids but also found in bacterial chromosomes. Each resistance gene in an integron is encoded in a mobile gene cassette that can be excised and then incorporated into another integron on another genome. Multiple cassettes with different resistance genes are commonly lined up, one after another, in an integron and expressed as a group from one upstream promoter. In some species of bacteria, such as Bacteroides, a chromosomal resistance gene may be within a conjugative transposon. The conjugative transposon may be excised to form an intermediate that may transfer and regenerate a double-stranded circle in another bacteria cell and integrate into its chromosome.
_
Examples of transmission of genetic material for AMR between microorganisms:
Genetic material is transferred between microorganisms through three main routes: (i) transformation—examples include recombination of foreign DNA from Streptococcus mitis to Streptococcus pneumoniae, conferring penicillin resistance via the formation of mosaic genes, and Neisseria gonorrhoeae where a mosaic penA gene is associated with ceftriaxone resistance; (ii) transduction—where antimicrobial resistance genetic material has been identified in phage DNA isolated from waste water treatment, and both extended-spectrum β-lactamase genes and mecA genes (the latter responsible for meticillin resistance in Staphylococcus aureus) have been identified in bacteriophage extracted from faecal samples at farms and abattoirs; and (iii) conjugation—with plasmids being responsible for example for the global dissemination of genes encoding carbapenemases such as New Delhi metallo-β-lactamase, as well as ESβLs. Furthermore integrative chromosomal elements (ICEs) can transfer these resistance genes between plasmids and the bacterial host chromosome in a range of Gram-negative species and streptococci. The combined effects of fast growth rates to large densities of cells, genetic processes of mutation and selection, and the ability to exchange genes, account for the extraordinary rates of adaptation and evolution that can be observed in the bacteria. For these reasons bacterial adaptation (resistance) to the antibiotic environment seems to take place very rapidly in evolutionary time. Bacteria evolve fast!
_____
Intrinsic Resistance:
Intrinsic resistance refers to the existence of genes in bacterial genomes that could generate a resistance phenotype, i.e., proto- or quasi-resistance. Different genera, species, strains, etc., exhibit ranges of antibiotic response phenotypes. The intrinsic resistome is an evolutionary ancient phenotype and can be defined as the intrinsic resistance of any bacterial species that has not been acquired as a result of exposure to antibiotics. Intrinsic resistance is usually the result of the reduced permeability of the bacterial envelope and the activity of multidrug efflux pumps. This suggests that the main physiological role of the components of intrinsic resistance involves the prevention of influx of toxic components by restricting the permeability of the cell or the active export of toxic compounds or their metabolites out of the cell. For example, an organism lacks a transport system for an antibiotic; or an organism lacks the target of the antibiotic molecule; or, as in the case of Gram-negative bacteria, the cell wall is covered with an outer membrane that establishes a permeability barrier against the antibiotic. Intrinsic resistance is the innate ability of a bacterial species to resist activity of a particular antimicrobial agent through its inherent structural or functional characteristics, which allow tolerance of a particular drug or antimicrobial class. This can also be called “insensitivity” since it occurs in organisms that have never been susceptible to that particular drug. Such natural insensitivity can be due to:
•lack of affinity of the drug for the bacterial target
•inaccessibility of the drug into the bacterial cell
•extrusion of the drug by chromosomally encoded active exporters
•innate production of enzymes that inactivate the drug
_
Examples of intrinsic resistance and their respective mechanisms:
| ORGANISMS | NATURAL RESISTANCE AGAINST: | MECHANISM |
| Anaerobic bacteria | Aminoglycosides | Lack of oxidative metabolism to drive uptake of aminoglycosides |
| Aerobic bacteria | Metronidazole | Inability to anaerobically reduce drug to its active form |
| Gram-positive bacteria | Aztreonam (a beta-lactam) | Lack of penicillin binding proteins (PBPs) that bind and are inhibited by this beta lactam antibiotic |
| Gram-negative bacteria | Vancomycin | Lack of uptake resulting from inability of vancomycin to penetrate outer membrane |
| Klebsiella spp. | Ampicillin (a beta-lactam) | Production of enzymes (beta-lactamases) that destroy ampicillin before the drug can reach the PBP targets |
| Stenotrophomonas. maltophila | Imipenem (a beta-lactam) | Production of enzymes (beta lactamases) that destroy imipenem before the drug can reach the PBP targets. |
| Lactobacilli and Leuconostoc | Vancomycin | Lack of appropriate cell wall precursor target to allow vancomycin to bind and inhibit cell wall synthesis |
| Pseudomonas aeruginosa | Sulfonamides, trimethoprim, tetracycline, or chloramphenicol | Lack of uptake resulting from inability of antibiotics to achieve effective intracellular concentrations |
| Enterococci | Aminoglycosides | Lack of sufficient oxidative metabolism to drive uptake of aminoglycosides |
| All cephalosporins | Lack of PBPs that effectively bind and are inhibited by these beta lactam antibiotics |
__
Clinical implications: Intrinsic Resistance:
Knowledge of the intrinsic resistance of a pathogen of concern is important in practice to avoid inappropriate and ineffective therapies. For bacterial pathogens which are naturally insensitive to a large number of classes of antimicrobials, such as Mycobacterium tuberculosis and Pseudomonas aeruginosa, this consideration can pose a limitation in the range of options for treatment and thus consequently further increase the risk for emergence of acquired resistance.
_______
Acquired resistance:
Several mechanisms are developed by bacteria in order to acquire resistance to antibiotics. All require either the modification of existing genetic material or the acquisition of new genetic material from another source. Acquired resistance is said to occur when a particular microorganism obtains the ability to resist the activity of a particular antimicrobial agent to which it was previously susceptible. Acquired resistance results from mutation or horizontal gene transfer via transformation, transduction or conjugation. Changes in bacterial genome through mutation or horizontal gene acquisition may consequently lead to a change in the nature of proteins expressed by the organism. Such change may lead to an alteration in the structural and functional features of the bacteria involved, which may result in changes leading to resistance against a particular antibiotic. Unlike intrinsic resistance, traits associated with acquired resistance are found only in some strains or subpopulations of each particular bacterial species. Laboratory methods are therefore needed to detect acquired resistance in bacterial species that are not intrinsically resistant. These same methods are used for monitoring rates of acquired resistance as a means of combating the emergence and spread of acquired resistance traits in pathogenic and non-pathogenic bacterial species. Because resistance traits are not naturally eliminated or reversed, resistance to a variety of antibiotics may be accumulated over time. This can lead to strains with multiple drug resistance, which are more difficult to kill due to reduced treatment options. Some antimicrobial resistance is brought about by multiple changes in the bacterial genome. For example, Isoniazid resistance of Mycobacterium tuberculosis results from changes in the following genes: katG gene which encodes a catalase; inhA gene which is the target for isoniazid; the oxyR gene and neighboring aphC gene and their intergenic region.
_
| ACQUIRED RESISTANCE THROUGH: | RESISTANCE OBSERVED | MECHANISM INVOLVED |
| Mutations | Mycobacterium tuberculosis resistance to rifamycins | Point mutations in the rifampin-binding region of rpoB |
| Resistance of many clinical isolates to fluoroquinolones | Predominantly mutation of the quinolone-resistance-determining-regiont (QRDR) of GyrA and ParC/GrlA | |
| E.coli, Hemophilius influenzae resistance to trimethoprim | Mutations in the chromosomal gene specifying dihydrofolate reductase | |
| Horizontal gene transfer | Staphylococcus aureus resistance to methicillin (MRSA) | Via acquisition of mecA genes which is on a mobile genetic element called “staphylococcal cassette chromosome” (SCCmec) which codes for penicllin binding proteins (PBPs) that are not sensitive to ß-lactam inhibition |
| Resistance of many pathogenic bacteria against sulfonamides | Mediated by the horizontal transfer of foreign folP genes or parts of it | |
| Enterococcus faecium and E. faecalis resistance to vancomycin | Via acquisition of one of two related gene clusters VanA and Van B, which code for enzymes that modify peptidoglycan precursor, reducing affinity to vancomycin. |
__
Biochemical mechanisms of acquired resistance:
Once bacteria acquire resistance gene through mutation or horizontal gene transfer, it employs various biochemical mechanisms to confer antibiotic resistance. Different classes of antibiotics possess specific modes of action by which they inhibit the growth or kill bacteria. These include inhibition of bacteria cell wall synthesis, inhibition of protein synthesis, inhibition of DNA synthesis, inhibition of RNA synthesis, competitive inhibition of folic acid synthesis and membrane disorganization. In all cases these effects involve the binding of antibiotics to specific bacterial molecular targets such as enzymes or the organelles. Bacteria can thus become resistant by developing mechanisms to prevent antibiotics binding to their molecular target. The four main methods by which bacteria achieve this are: inactivating or degrading antibiotics, modifying the target site, decreasing cell wall permeability (reducing antibiotic entry into bacterial cells) or active efflux, and metabolic bypass. Bacteria often possess multiple resistance mechanisms making them resistant to several classes of antibacterial agents.
_
The four main mechanisms by which microorganisms exhibit resistance to antimicrobials are:
_
Strategy 1: Preventing access:
Antimicrobial compounds almost always require access into the bacterial cell to reach their target site where they can interfere with the normal function of the bacterial organism. Porin channels are the passageways by which these antibiotics would normally cross the bacterial outer membrane. Some bacteria protect themselves by prohibiting these antimicrobial compounds from entering past their cell walls. For example, a variety of Gram-negative bacteria reduce the uptake of certain antibiotics, such as aminoglycosides and beta lactams, by modifying the cell membrane porin channel frequency, size, and selectivity. Prohibiting entry in this manner will prevent these antimicrobials from reaching their intended targets that, for aminoglycosides and beta lactams, are the ribosomes and the penicillin-binding proteins (PBPs), respectively.
This strategy has been observed in:
•Pseudomonas aeruginosa against imipenem (a beta-lactam antibiotic)
•Enterobacter aerogenes and Klebsiella spp. against imipenem
•Vancomycin intermediate-resistant S. aureus or VISA strains with thickened cell wall trapping vancomycin
•Many Gram-negative bacteria against aminoglycosides
•Many Gram-negative bacteria against quinolones
_
Strategy 2: Eliminating antimicrobial agents from the cell with expulsion via efflux pumps.
To be effective, antimicrobial agents must also be present at a sufficiently high concentration within the bacterial cell. Some bacteria possess membrane proteins that act as an export or efflux pump for certain antimicrobials, extruding the antibiotic out of the cell as fast as it can enter. This results in low intracellular concentrations that are insufficient to elicit an effect. Some efflux pumps selectively extrude specific antibiotics such as macrolides, lincosamides, streptogramins and tetracyclines, whereas others (referred to as multiple drug resistance pumps) expel a variety of structurally diverse anti-infectives with different modes of action.
This strategy has been observed in:
•E.coli and other Enterobacteriaceae against tetracyclines
•Enterobacteriaceae against chloramphenicol
•Staphylococci against macrolides and streptogramins
•Staphylococcus aureus and Streptococcus pneumoniae against fluoroquinolones
These efflux pumps are variants of membrane pumps possessed by all bacteria, both pathogenic and non-pathogenic, to move lipophilic or amphipathic molecules in and out of the cells. Some are used by antibiotic producers to pump antibiotics out of the cells as fast as they are made, and so constitute an immunity protective mechanism for the bacteria to prevent being killed by their own chemical weapons.
_
Strategy 3: Inactivation of antimicrobial agents via modification or degradation:
Another means by which bacteria preserve themselves is by destroying the active component of the antimicrobial agent. A classic example is the hydrolytic deactivation of the beta-lactam ring in penicillins and cephalosporins by the bacterial enzyme called beta lactamase. The inactivated penicilloic acid will then be ineffective in binding to PBPs (penicllin binding proteins), thereby protecting the process of cell wall synthesis.
This strategy has also been observed in:
Enterobacteriaceae against chloramphenicol (acetylation)
Gram negative and Gram positive bacteria against aminoglycosides (phosphorylation, adenylation, and acetylation)
_
Strategy 4: Modification of the antimicrobial target:
Some resistant bacteria evade antimicrobials by reprogramming or camouflaging critical target sites to avoid recognition. Therefore, in spite of the presence of an intact and active antimicrobial compound, no subsequent binding or inhibition will take place.
This strategy has been observed in:
•Staphylococci against methicillin and other beta-lactams (Changes or acquisition of different PBPs that do not sufficiently bind beta-lactams to inhibit cell wall synthesis.)
•Enterococci against vancomycin (alteration in cell wall precursor components to decrease binding of vancomycin)
•Mycobacterium spp. against streptomycin (modification of ribosomal proteins or of 16s rRNA)
•Mutations in RNA polymerase resulting in resistance to the rifamycins
•Mutations in DNA gyrase resulting in resistance to quinolones
_
_
Quorum sensing (QS):
Bacterial cell-to-cell communication is called quorum sensing. The communication is possible because bacteria can produce, release and detect chemical signals – called signalling molecules. If there are few bacteria in the area, the concentration of signalling molecules is low, so certain genes are prevented from being expressed and the bacteria divide as much as possible. However, once there are enough bacteria in the local area – these signalling molecules cause the bacteria to produce new proteins that allow more effective infection of human cells. Microbes communicate with each other and exchange signalling chemicals (Autoinducers). These autoinducers allow bacterial population to coordinate gene expression for virulence, conjugation, apoptosis, mobility and resistance. Single autoinducer from single microbe is incapable of inducing any such change. But when its colony reaches a critical density (quorum), threshold of autoinduction is reached and gene expression starts. QS signal molecules AHL, AIP, AI-2 & AI-3 have been identified in gram-ve bacteria. AI-2 QS –system is shared by gram +ve bacteria also. QS inhibitors have been synthesized and have been isolated from several natural extracts such as garlic extract. QS inhibitors have shown to be potent virulence inhibitor both in in-vitro and in-vivo, using infection animal models. New treatments involving disrupting this communication – with many pharmaceutical companies developing new chemicals that directly inhibit the signal production and signal reception – so the bacteria can be more effectively killed by the host immune system. This new concept is highly attractive because the spread of resistance seems highly unlikely. Resistant bacteria would not receive any signals from other bacteria as it sends and receives different signals. This would mean the resistant bacteria would not be able to co-ordinate fundamental processes of infection, such as biofilm formation and protein production that inhibit our immune response.
____
Biological versus Clinical Resistance:
Biological resistance refers to changes that result in the organism being less susceptible to a particular antimicrobial agent than has been previously observed. When antimicrobial susceptibility has been lost to such an extent that the drug is no longer effective for clinical use, the organism is then said to have achieved clinical resistance. It is important to note that often, biologic resistance and clinical resistance do not necessarily coincide. From a clinical laboratory and public health perspective it is important to realize that biologic development of antimicrobial resistance is an ongoing process, while clinical resistance is dependent on current laboratory methods and established cut-offs. Our inability to reliably detect all these processes with current laboratory procedures and criteria should not be perceived as evidence that they are not occurring.
_
Direct and indirect development of AMR:
Direct development of resistance:
Selection pressure exerted by antibiotics over bacteria resulting in development of AMR is direct effect of antimicrobial use.
Indirect Effects on Resistance:
For any infectious disease, the infection or colonization status of any one (index) patient affects the risk of infection or colonization of others. Measures (such as vaccination or antibiotic treatment) that change the incidence or duration of infection in one person will affect that person’s contacts. Just as vaccination programs benefit those who are not vaccinated because of the phenomenon of herd immunity, antibiotic usage by some persons may increase the risk of colonization or infection with resistant organisms in people who have not received antibiotics. Members of a population experience indirect effects of antimicrobial use, defined as the enhancement of risk for acquiring a resistant organism, because of the use of antimicrobials by other persons in the group or population. The relationship between antibiotic usage and antibiotic resistance for many types of pathogens is largely mediated by indirect effects or population-level selection. When resistant and susceptible organisms compete to colonize or infect hosts, and use of an antibiotic has a greater impact on the transmission of susceptible bacteria than resistant ones, then increasing use of the antibiotic will result in an increase in frequency of organisms resistant to that drug in the population, even if the risk for treated patients is modest. Antimicrobial use and patient-to-patient transmission are not independent pathways for promoting of antimicrobial resistance, rather they are inextricably linked.
Figure above shows four mechanisms by which antibiotic treatment can create selection for resistance in the population, showing direct effects—increased resistance in treated (yellow) vs. untreated (white) hosts, and indirect effects—increased resistance in others (turquoise) due to treatment of specific hosts.
(A) Subpopulations (usually mutants) of resistant (red) bacteria are present in a host infected with a predominantly susceptible (green) strain; treatment fails, resulting in outgrowth of the resistant subpopulation, which can then be transmitted to other, susceptible hosts (turquoise).
(B) Successful treatment of an individual infected with a susceptible strain reduces the ability of that host to transmit the infection to other susceptible hosts, making those hosts more likely to be infected by resistant pathogens than they would otherwise have been, and shifting the competitive balance toward resistant infections.
(C) Treatment of an infection eradicates a population of susceptible bacteria carried (often commensally) by the host, making that host more susceptible to acquisition of a new strain. If the newly acquired strain has a high probability of being resistant (as in the context of an outbreak of a resistant strain), this can significantly increase the treated individual’s risk of carrying a resistant strain, relative to an untreated one.
(D) Treatment of an infection in an individual who is already colonized (commensally) with resistant organisms may result in increased load of those organisms if competing flora (perhaps of another species) are inhibited—leading to increased shedding of the resistant organism and possibly to increased individual risk of infection with the resistant organism.
______
______
β-lactamase:
Enzymes that catalyze the cleavage of β-lactam rings in penicillins, cephalosporins, monobactams, and carbapenems were first described by Abraham and Chain in 1940. These enzymes confer resistance to β-lactam antibiotics on bacteria that produce them. β-lactamases are ancient, theorized to have evolved 1–2 billion years ago, but the emergence and spread of penicillin-resistant staphylococci in hospitals in the 1950s showed how penicillin use could select producers from a population of nonproducers. Beta-lactamases (β-lactamases) are enzymes produced by bacteria (also known as penicillinase) that provide multi-resistance to β-lactam antibiotics. Beta-lactamase provides antibiotic resistance by breaking the antibiotics’ structure. These antibiotics all have a common element in their molecular structure: a four-atom ring known as a β-lactam. Through hydrolysis, the lactamase enzyme breaks the β-lactam ring open, deactivating the molecule’s antibacterial properties. Beta-lactam antibiotics are typically used to treat a broad spectrum of Gram-positive and Gram-negative bacteria. Beta-lactamases produced by Gram-negative organisms are usually secreted, especially when antibiotics are present in the environment. Carbapenemases are β-lactamases with versatile hydrolytic capacities. They have the ability to hydrolyse penicillins, cephalosporins, monobactams, and carbapenems. Bacteria producing these β-lactamases may cause serious infections in which the carbapenemase activity renders many β-lactams ineffective. Gram positive bacteria have also become resistant to b-lactam antibiotics due variations in penicillin-binding proteins that lead to reduced binding to the beta lactam and other mechanisms like altered target and decreased uptake although few of them do generate b-lactamase.
_
_
Classification of β-lactamase:
Two classification schemes for β-lactamases are currently in use. The molecular classification is based on the amino acid sequence and divides β-lactamases into class A, C, and D enzymes which utilize serine for β-lactam hydrolysis and class B metalloenzymes which require divalent zinc ions for substrate hydrolysis. The functional classification takes into account substrate and inhibitor profiles in an attempt to group the enzymes in ways that can be correlated with their phenotype in clinical isolates. Major groupings generally correlate with the more broadly based molecular classification. The updated system includes group 1 (class C) cephalosporinases; group 2 (classes A and D) broad-spectrum, inhibitor-resistant, and extended-spectrum β-lactamases and serine carbapenemases; and group 3 metallo-β-lactamases. Several new subgroups of each of the major groups are described, based on specific attributes of individual enzymes. A list of attributes is also suggested for the description of a new β-lactamase, including the requisite microbiological properties, substrate and inhibitor profiles, and molecular sequence data that provide an adequate characterization for a new β-lactam-hydrolyzing enzyme.
___
___
The figure above shows number of mechanisms used by common antibiotics to deal with bacteria and ways by which bacteria become resistant to them.
____
The molecular mechanisms of resistance to antibiotics have been studied extensively and have involved investigations of the genetics and biochemistry of many different facets of bacterial cell function. In fact, the study of antibiotic action and resistance has contributed significantly to our knowledge of cell structure and function. Resistance processes are widely distributed in the microbial kingdom and have been well described for a variety of commensals and pathogens; most can be disseminated by one or more distinct gene transfer mechanisms.
_
Modes of action and resistance mechanisms of commonly used antibiotics:
__________
Novel antibiotic resistance mechanism:
Superbugs deploy ‘decoy flares’ like fighter jets to avoid being killed by antibiotics: a 2016 study:
An MRSA bacterial cell releases decoys to evade an antibiotic. Scientists have discovered an astonishing new strategy deployed by superbugs in their evolutionary struggle with antibiotics – decoy ‘flares’ which distract the drugs that would otherwise kill them. About a third of MRSA bacteria, which kill thousands of people around the world, are largely immune to the current antibiotic of last resort. In a new academic paper in the journal Nature Microbiology, researchers said they had found out a reason why. When it comes under attack, the bug can release ‘decoys’ which the scientists compared to the flares used by warplanes to distract heat-seeking missiles. They are made from the same type of fat that makes up the outer layer of the superbug’s cells. The antibiotic, daptomycin, normally latches onto this fat, drills a hole through the cell wall and kills the bacteria. But faced with a sea of fat decoys, they attack those instead and are used up. These fat molecules act in a similar way to the decoy flares released by fighter planes to avoid a missile. The antibiotic mistakenly targets the decoys, allowing the bacteria to evade destruction. This is the first time this decoy system has been seen in MRSA. A similar decoy mechanism has been found in E. coli, leading the researchers to believe this may be a defence mechanism used by “many other bacteria”. Further work led to the discovery that another antibiotic, called oxacillin, can partially prevent the release of the fatty decoys. It is thought that even though a strain of MRSA bacteria might be resistant to both daptomycin and oxacillin, using the two drugs together could be an effective strategy. A team in Australia is currently carrying out a clinical trial of this technique. And a next generation antibiotic, also in clinical trials, has shown signs of being able to stop the production of the decoys.
_______
Mechanism of Spread of Resistance:
A resistant strain made prevalent by selection in the bacterial populations of one host is more likely to be among the strains that the host transfers to a second host. Similar selection in the second host would boost the strain’s chances of becoming established, amplified, and then transferred to a third host. These considerations would predict that resistant strains travel the world selectively through networks of hosts being treated with antimicrobial agents. The experiences of intensive care units, day care centers, and feedlots tend to confirm this prediction. Relieved of competition from susceptible strains, resistant strains spreading through networks of antimicrobial-treated hosts would compete more directly with one another. The history of antimicrobial resistance has often been that of a successful resistance construct evolving under selection somewhere, emerging under further selection, and then spreading nearly everywhere. Strains of Staphylococcus aureus belonging to a few phage types that possessed an inducible penicillinase gradually spread throughout the world’s hospitals in the 1950s and throughout communities everywhere in the 1960s. A few clones with intricate constructs expressing the mec gene then spread methicillin resistance through the world’s hospitals. In the 1970s, a single plasmid carried gentamicin resistance to several genera of enteric bacteria in a number of hospitals in different parts of the United States and in one hospital in Venezuela, none of which had seen any gentamicin-resistant enteric bacteria until then. A particular transposon, Tn1331, was first noted to encode amikacin resistance in Argentina and Chile but was later seen in other parts of the world. Resistance to sulfonamides, possibly the most prevalent type of resistance, has been found throughout the world encoded by only 2 resistance genes. One of them was found to be virtually ubiquitous on a small multicopy plasmid also carrying a streptomycin-resistance gene; this may contribute to the persistence of resistance to streptomycin decades after its use in human therapy has almost completely ceased. Several multidrug-resistant plasmids found to be endemic in several Salmonella serotypes isolated from animals in some US states were found in clinical isolates of the same serotypes from humans infected in distant states. After decades of penicillin use, certain strains of Streptococcus pneumoniae acquired long, contiguous nucleotide sequences expressing foreign penicillin-binding proteins that made the strains resistant to penicillin. Once arisen, these new chromosomal genetic constructs spread clonally in strains of pneumococci belonging to serotypes that could be traced through countries and continents. One such construct in serotype 6B, originally prevalent in Spain, was later found in other countries in the Western Hemisphere; its eventual incursion into Iceland accounted for nearly all penicillin-resistant pneumococci there.
________
Mobile resistome:
Certain antibiotic resistance genes are easily transferred from one bacterial species to another, and can move between farm animals and the human gut. A team led by Chinese researchers has characterized this “mobile resistome,” which they say is largely to blame for the spread of antibiotic resistance. They found that many antibiotic resistance genes that are shared between the human and animal gut microbiome are also present in multiple human pathogens. These findings are published September 9, 2016 in Applied and Environmental Microbiology, a journal of the American Society for Microbiology. The so-called “transfer network” of antibiotic resistance genes described in the paper is very forward reaching and will have great impact not only on our understanding of this modern microbial dilemma but also on how human healthcare agencies and research institutes attempt to cope with it. The network of horizontal gene transfer is shaped largely by phylogeny and ecological constraints. That is, resistance gene transfers from one species of bacteria to another are more common within the same phylum than between different phyla, and more common within a single microbiome than between microbiomes. On the latter point, the investigators wrote that successful gene transfer requires contact between donor and recipient. The recent mobile resistance gene transfer that has taken place between livestock and human gut microbiomes is especially important for policy-makers. Much of the resistance in farm animals is generated by feeding them large quantities of antibiotics, which is done because it encourages the animals to grow faster. One consideration, from the worldwide ecological view, is that bacteria of animal origin may face more antibiotic selection pressure because more antibiotics (nearly 80 percent in the United states) are consumed by animals as growth-promotors, infection prevention, and clinical treatments. The high exchange frequency of mobile [antibiotic resistance genes] between animals and humans or environmental bacteria is also noteworthy. Resistance genes can also be transferred between organisms via mobile genetic elements (MGEs) such as plasmids, and transferable resistance is often more important clinically in MDR Gram-negatives than is resistance arising from mutation. There is ample evidence that MGEs are able to transfer resistance mechanisms between genera; for example, Hegstad et al describe MGEs of enterococci being transferred to Staphylococcus aureus. This ability of bacteria to transfer resistance mechanisms provides a major challenge to preventing the emergence of resistance. The evidence suggests that antibiotic resistance genes in human bacterial pathogens originate from a multitude of bacterial sources, indicating that the genomes of all bacteria can be considered as a single global gene pool into which most, if not all, bacteria can dip for genes necessary for survival. In terms of antibiotic resistance, plasmids serve a central role, as the vehicles for resistance gene capture and their subsequent dissemination.
_
Environmental antibiotic resistome:
It has been known for some time that bacterial strains resistant to antibiotics can be isolated by plating environmental bacteria on antibiotic-containing media in the laboratory. This is not surprising for antibiotic-producing actinomycetes, since most possess genes encoding resistance to the compounds that they produce. In several cases, the resistance mechanisms have been identified and shown to be specific enzymatic modifications of the antibiotics. Streptomycetes have long been known to produce a variety of β-lactamases that may well be the source of some of the clinical forms of β-lactam resistance. Environmental Kluyvera species have been found to be the origins of the CTX-M genes. In other cases, resistance of producing organisms to their products has been identified as due to efflux systems. Multiple mechanisms of resistance, as found in the tetracycline producer Streptomyces rimosus, are frequent in producing bacteria. Based on biochemical and genetic similarities, such resistance mechanisms have presaged those found subsequently in antibiotic-resistant pathogens. In a recent, all-inclusive approach to quantifying the r genes/phenotype density in the environment, Wright and colleagues screened a collection of morphologically distinct spore-forming actinomycetes (including many known antibiotic-producing strains) for resistance to 21 different antibiotics. A significant number of strains were resistant to an average of 7 or 8 antibiotics; they were naturally multidrug resistant. The population of r genes in nature is referred to as the environmental antibiotic resistome. Clearly, different environments would be expected to vary in the number and type of resistances. Novel resistance mechanisms, as well as many mechanisms related to those found in pathogens, were identified in the collection. This is the best evidence available for the presence of a vast environmental pool of genes with the potential to be captured and expressed as resistance determinants for any overused inhibitor. However, more studies are necessary to establish a strong environment-clinic connection. Similar surveys of other antibiotic-producing bacteria, such as the Bacillaceae, pseudomonads, cyanobacteria, and the extensive family of Actinobacteria, a phylogenetic group known to produce many low-molecular-weight molecules, will be valuable in extending our understanding of the nature of r genes existing in the wild.
_
The Subsistome:
Dantas and coworkers have taken a complementary approach to that of D’Costa et al. by screening soil bacteria for biochemical processes that degrade or inactivate antibiotics. Hundreds of strains were randomly isolated from 11 diverse urban and rural soils and tested for the ability to subsist or grow on one or more of 18 different antibiotics as sole carbon and nitrogen sources. Perhaps surprisingly, many strains were isolated that grew efficiently on common antimicrobials, including aminoglycosides, fluoroquinolones, and other classes. Most of the strains identified in this study were proteobacteria, and more than 40% were Burkholderia spp.; pseudomonads were also well represented. Obviously, catabolic pathways responsible for antibiotic digestion in nature provide a rich source of potential resistance determinants; additional studies should reveal novel mechanisms of resistance to most antibiotic classes. Work on antibiotic-catabolizing bacteria was reported in the 1970s, but the studies of Dantas and colleagues have exposed the full extent and distribution of degradation/r genes in the environment and further verified the roles played by reservoirs of soil bacteria as origins of antibiotic r genes.
______
The Parvome (the World of Small Molecules):
There is lamentable ignorance of the roles of many millions of low-molecular-weight organic compounds that are produced by bacteria, other microbes, and plants. Their production requires well-defined biochemical pathways and involves biosynthetic gene clusters that are frequently larger than 100 kb. The study of the myriad aspects of small-molecule biology deserves attention. As a collective noun for the infinite world of bioactive small molecules (usually less than 3,000 Da) produced by living organisms, authors have coined the word “parvome,” a combination of parv- (Greek prefix: small) and -ome (Latin suffix: group).What are the origins of small bioactive organic molecules such as antibiotics? What are the natural roles of these compounds? An even more intriguing question concerns the evolution of the complex biosynthetic pathways of all the bioactive compounds produced in nature. The structural components of antibiotics appear to have existed in the biosphere for billions of years, as evidenced by the number of primordial amino acid derivatives (many of them components of nonribosomal peptides) found in meteorites and by products from “prebiotic” reaction conditions. Baltz has calculated that the biosynthetic pathway for a polyketide molecule such as erythromycin may have evolved as many as 800 million years ago, and the streptomycin biosynthetic pathway is at least 600 million years old.
_
Phylogenetic analysis shows that the OXA b-lactamase genes have been on Plasmids for millions of years: a 2002 study:
The OXA genes encode a class of b-lactamases that confer resistance to a wide range of b-lactam antibiotics. To determine whether the diversity of the OXA b-lactamases is the result of recent or ancient events, and to determine whether mobilization of the OXA genes from chromosomes to plasmids occurred recently or long ago, authors have constructed a Bayesian phylogeny of the OXA b-lactamase genes. Analysis of that phylogeny shows that much of the diversity is the result of ancient events and that the OXA genes were mobilized from chromosomes to plasmids on at least two independent occasions that occurred millions of years ago. That observation contradicts the commonly held impression that mobilization of antibiotic resistance genes is strictly the result of modern use of antibiotics.
Note:
This study does support my hypothesis that resistance genes can be mobilized from chromosomes to plasmids to be transferred to other bacteria.
______
______
Commensals and AMR:
The role of commensal bacteria in the spread of antibiotic resistance is recognized as a vital component in understanding how to preserve the power of antibiotics. Excepting tuberculosis, the majority of bacterially caused illness results from infection by commensal organisms, such as Escherichia coli, streptococci, and staphylococci. Thus, better knowledge of the biology of antibiotic resistance in commensal bacteria is a crucial component to treating these life-threatening diseases. The two roles that commensal bacteria play are as reservoirs of antibiotic resistance genes and as drug-resistant opportunistic pathogens. In the United States, roughly 75% of clinical isolates are commensal organisms that can also exist as opportunistic pathogens (E. coli, Enterococcus, Staphylococcus, and Streptococcus). As serious as the commensal problem is in the developed world, it is far more serious in the developing world. Forty-six percent of childhood mortality is due to diarrhea and pneumonia, much of which is bacterial in cause. Most of these disease-causing bacteria are either related to commensals or are themselves commensals in other organisms or even in humans. Thus, better knowledge of the biology of antibiotic resistance in commensal bacteria is a crucial component to treating these life-threatening diseases.
_
Defining Commensals:
One of the most conceptually difficult terms to define in microbiology is commensal. Many species of bacteria contain members that can coexist with human hosts without causing disease, as well as members which cause disease. To complicate matters further, many strains within a species are able to switch between pathogenic and commensal life histories, depending on the ecological context. For example, at a global level, E. coli strains belonging to the ST69 complex account for roughly 7% of E. coli isolated from humans. This clonal complex is also capable of behaving as a highly virulent pathogen when it gains access to the urinary tract because it possesses specific adaptations for survival and reproduction in that habitat. This clone appears to be well adapted to commensal and pathogenic strategies. Additionally, many strains are virulent in some hosts and not others. Shiga toxin-producing E. coli cause severe gastrointestinal illness in humans, but in cattle unable to cause disease. Even within the same host species, a given strain can vary in its ability to cause disease, as is the case with travellers’ diarrhoea, which sickens naive, unexposed hosts. One useful distinction is to distinguish between ‘‘pathogenic commensals’’ that can cause disease when a patient is vulnerable or the bacterium gains access to a sterile site and ‘‘non-pathogenic commensals’’ which are incapable of causing disease, such as some lactobacilli. Another distinction is to classify bacteria based on the source of isolation. Commensals can be defined as those isolates collected from asymptomatic individuals and from ‘‘typical’’ colonization sites, such as E. coli from feces and Streptococcus from nasal passages, or those isolated from environmental or non-human animal sources. A working definition of commensal is an organism which typically does not cause disease, although some individual strains within a commensal species may be able to cause disease. A commensal isolate is defined as an isolate which has not been acquired from a diseased animal or person, even though that isolate could cause disease in an atypical site, and which would not cause disease in humans following typical colonization. Under this definition, an E. coli isolate that has the capacity to be a virulent bloodstream pathogen would be considered a commensal since the bloodstream is not the primary habitat of E. coli, whereas a Shiga toxin-producing E. coli isolate would not be considered a commensal, even though it does not cause disease in bovines, because this isolate will cause disease following intestinal colonization in humans.
_
Commensals as Reservoirs of Antibiotic Resistance:
As discussed above, the meaning of the phrase ‘‘commensal organism’’ has been a subject of much debate, and there has been little resolution of this issue. Most host-associated organisms fall along a continuum of pathogenicity. Some, such as the Shigella complexes of E. coli and diarrheagenic E. coli, are essentially pathogens. Others, such as the extraintestinal pathogenic E. coli (e.g., urinary tract infections) and the vancomycin-resistant enterococci, are commensals in their usual habitat (the intestinal tract), but when present in non-commensal sites are opportunistic pathogens. In opportunistic pathogens, virulence will be dependent on the complement of virulence-associated loci. Consequently, commensalism should be viewed as a sliding scale or a continuum, and not as a binary state. When commensalism is viewed as a continuum and not an ‘‘either-or’’ state, there are two ways in which commensals can function as reservoirs of antibiotic resistance for pathogens:
1. Commensals may act as a potential source of antibiotic resistance genes that can be transferred to pathogens via horizontal gene transfer.
2. Resistant commensals which are able to behave as opportunistic pathogens will increase in frequency due to selection for resistance. This would increase the likelihood that a given opportunistic infection would be antibiotic resistant and thus difficult to treat.
These are not mutually exclusive roles. As the proportion of resistant commensals increases, the likelihood that successful gene transfer will occur also increases via two mechanisms. First, if pre-existing resistant clones are favored, then that environment will also favor resistant transferrants. Second, as resistant commensals increase in number, there are more potential resistant donors. Many examples of horizontal transfer of antibiotic resistance genes from commensals to pathogens have been documented since the 1970s. Given widespread antibiotic resistance in many organisms, it is not clear, at this point, to what extent the further generation of novel resistance genotypes via horizontal gene transfer would be the primary response to selection versus the clonal spread of pre-existing resistance genotypes.
_
There have been very few systematic studies to investigate the acquired antibiotic resistance in lactic acid bacteria (LAB) of food origin. However, they are lately expanding due to increased interest in probiotic lactic acid bacteria and genetic modification of LAB. When LAB live in a biotope regularly challenged by antibiotics (human or animal intestine, bovine udder), the acquired antibiotic resistance is found in Enterococcus, Lactococcus and Lactobacillus species. The resistant bacteria may interact with the resident human microflora and possibly transfer or acquire antibiotic resistance determinants by horizontal gene transfer. Large numbers of probiotic bacteria are consumed to maintain and restore the microbial balance in the intestines. It must be kept in mind that they have a potential to transfer antibiotic resistances to pathogenic bacteria. For these and other applications, the safety aspects of these bacteria are of concern, including the presence of potentially transferable antibiotic resistances. Bacteria that normally reside in the human colon can transfer resistance genes among themselves. This type of transfer becomes a huge problem when these harmless commensal bacteria transform into pathogens. The environment is replete with drug resistance genes, among both pathogen and commensal bacteria. Once acquired, resistance genes are not easily lost. Instead, they become a relatively stable part of a genome. Additional resistance determinants may join those already prevailing, thus broadening the multidrug resistance phenotype and further diminishing treatment options. An increasing number of bacterial isolates is resistant to practically all available therapeutic agents. Multidrug resistance has been demonstrated in Escherichia coli, Salmonella enterica serovar Typhimurium, Shigella dysenteriae, Enterococcus faecium, Staphylococcus aureus, Mycobacterium tuberculosis, Haemophilus influenzae, Pseudomonas aeruginosa, Klebsiella pneumoniae, Acinetobacter baumannii, Stenotrophomonas maltophilia, Xanthomonas and Burkholderia. Thus, the emergence of antibiotic resistance in bacterial populations is a relevant field of study in molecular and evolutionary biology as well as in medical practice.
_
Hierarchy of commensals:
The figure above shows hierarchy of commensals (not to scale). This group of microbes comprises only a tiny fraction of the total microbial environment. Greatly underated and understudied are the multitudes of “core” and transient colonizers, i.e., commensals that constitute the major reservoirs of resistance genes. To some degree, commensals can be distinguished by their place in the environment and the relationships with their hosts. Some colonizers of the skin, oropharynx, and intestinal tract rarely if ever cause disease (e.g., the lactic acid bacteria). Yet another group is considered generally nonpathogenic, but when imbalances or shifts occur in the selective pressures on their microbial niches, these species can be propelled to the status of pathogens, made more problematic if they have acquired resistance or virulence genes from neighboring commensals. These constitute the group of opportunistic or “pathogenic commensals.” Staphylococcus aureus, which regularly or transiently colonizes about 80% of humans, now frequently crosses this border. S. epidermidis and other traditionally commensal coagulase-negative staphylococci only occasionally appear as nosocomial infections and only under extreme selective pressures, such as indwelling catheters and depressed immunologic states. Species that commonly harbor native or “constitutive” resistance in their chromosomes (e.g., Pseudomonas, Stenotrophomonas, and Acinetobacter), also may emerge under these conditions. Most commensals, however, exist as environmental residents of soil and water habitats, many of which may become transient colonizers of humans and animals through the food chain and other routes of exposure. The accumulated evidence suggests widespread gene exchange among these groups
_
A pivotal role for the gut microbiota:
The intestinal microbiota forms the main reservoir of multidrug-resistant GNB in critically ill patients. While similar data are currently not available for ICU-acquired infections, some studies have showed that high intestinal densities of resistant bacteria increase the risk of intestinal translocation, urinary tract infections and cross-transmission. Antibiotics that reach this microbiome promote the growth of resistant bacteria over the susceptible ones, and each daily dose may exert a significant impact in terms of selective pressure. This appears notably relevant for carbapenems, fluoroquinolones or cephalosporins with biliary excretion such as ceftriaxone. Extended treatment with colistin has also been shown to increase the likelihood of colonization with colistin-resistant GNB, including both mutants from otherwise colistin-susceptible species, and intrinsically colistin-resistant Enterobacteriaceae. The spectrum, duration of exposure and fecal concentration of the antibiotic may all play a role. Therefore, and although the ecological benefit of such an approach remains to be formally demonstrated, de-escalation to the antimicrobial regimen with the narrower spectrum and the lower intestinal excretion should be logically discussed when culture and susceptibility testing results become available. In this respect, new phenotypic and molecular diagnostic tools may fasten the detection of multidrug-resistant GNB—or rule them out precociously—thereby assisting ICU physicians for earlier adjustments of broad-spectrum empirical regimen.
_
Colonisation resistance:
Colonization resistance is the mechanism whereby the intestinal microflora protects itself against incursion by new and often harmful microorganisms. Colonization resistance was first identified in 1967 and was initially referred to as antibiotic associated susceptibility. It was observed that animals being treated with the antibiotic streptomycin were susceptible to Salmonella enterica at doses 10,000 fold lower than the standard minimal infectious dose. This led to investigations about the mechanisms utilized by endogenous microbial populations that conferred protection against exogenous pathogens attempting to colonize the gut flora. Recently it has been observed that colonization resistance can occur within the host in a ‘direct’ or ‘indirect’ manner. The former refers to particular components of the microbiota directly competing with exogenous pathogens for nutritional niches (e.g. Bacteroides thetaiotaomicron directly competes with Citrobacter rodentium for carbohydrates in the intestinal lumen) or by producing growth inhibitors (e.g. Bacteroides thuringiensis can secrete bacteriocin that directly targets spore-forming Clostridium difficile, thus inhibiting its growth through an unknown mechanism), that directly inhibit the colonizing pathogen. Indirect colonization resistance is thought to be mediated through the induction of immune responses in the host that concomitantly inhibit the colonizing pathogen. An example of this has been observed with B. thetaiotaomicron, which can induce the host to produce antimicrobial C-type lectins REGIIIγ and REGIIIβ, both anti-microbial peptides that target gram-positive bacteria.
_____
Resistant bacteria accumulate multiple resistance determinants:
The long-term use of a single antibiotic (that is, for more than 10 days) will select for bacteria that are resistant not only to that antibiotic, but to several others. This phenomenon was found to occur after the prolonged use of tetracycline for urinary tract infections and for acne. Under continued antimicrobial selection, the susceptible intestinal and/or skin flora may become colonized by organisms that are resistant not only to the ingested drug, but also to other, structurally unrelated drugs. In animals, MDR emerged after the application of sub-therapeutic (growth promotion) levels of tetracyclines in feed. Within days, chickens began excreting tetracycline-resistant E. coli; by two weeks, the excreted E. coli were resistant to several antibiotics. This phenomenon reflects the linkage of different resistance genes on the same transposon or plasmid. It is unclear, however, why multiple resistance plasmids eventually emerge with the prolonged use of a single antimicrobial. Bacteria that are already resistant to one growth-inhibitory agent seem to be favored in recruiting additional resistance traits from other bacteria sharing the environment: it was from the doubly resistant (penicillin and tetracycline) strains of N. gonorrhoeae that the new fluoroquinolone-resistant strains emerged.
_______
Loss of resistance is slow:
Resistant bacteria may rapidly appear in the host or environment after antibiotic use, but they are slow to be lost, even in the absence of the selecting antibiotic. This phenomenon reflects the minimal survival cost to the emerging resistant strains. In addition, resistance genes are often linked with genes specifying resistance to other antimicrobials or toxic substances on the same plasmids. The presence of MDR plasmids assures maintenance of the plasmid as long as any one of the resistances provides a survival advantage to the host bacterium. This principle also applies to determinants of resistance to biocides such as quaternary ammonium compounds, because biocide efflux genes can be found on plasmids bearing genes for resistance to antibiotics in S. aureus. Some studies have, however, tracked a decline in resistance frequencies when an antibiotic is removed. A significant countrywide reversal of macrolide resistance in S. pyogenes resulted from a Finnish nationwide campaign to reduce macrolide usage. In 2 years, resistance declined from about 20% to less than 10%. Nonetheless, resistance generally persists at some low level and reintroduction of the antimicrobial will reselect resistant strains despite months or even years of non-use. Replacement by susceptible flora represents a chief contribution to a decrease in resistant strains. For example, despite being put into clean cages, chickens previously fed tetracycline-laced feed were found to continue to excrete tetracycline-resistant E. coli at high frequencies. When placed in separate cages and moved to a new location in the barn every 2–3 days, however, the resistance frequency dropped. This ‘dilution’ of resistant strains was similarly accomplished by housing the chickens with greater numbers of cage mates that excreted susceptible flora. The findings suggest that the fastest way to eliminate resistant strains is to outnumber them with susceptible strains.
______
______
Diseases caused by resistant microorganisms:
_
List of Common Bacteria with High Antibiotic Resistance *
To have a better understanding of antibiotic resistance, the following table lists common bacteria that have become highly resistant, associated antibiotics with reduced activity, and antibiotics that may be appropriate for treatment of those resistant bacteria.
| Current and Emerging Resistant Bacteria | Type | Representative Clinical Infections | Antibiotics Associated with Resistance* | Treatment Options (as determined based on culture & sensitivity, local guidelines, clinical presentation) |
| Methicillin-resistant Staphylococcus aureus (MRSA) | gram (+) cocci | skin/soft tissue infections, UTI, bacteremia, toxic shock syndrome, pneumonia, osteomyelitis, endocarditis, meningitis; assoc. with IV catheters | beta-lactam antibiotics (eg., oxacillin, penicillin, nafcillin, amoxicillin, and most cepholosporins) erythromycin | Vancomycin– alternatives: linezolid; clindamycin (confirm with D-test); daptomycin; TMP-SMX; quinupristine-dalfopristin |
| Vancomycin inter- mediate and resistant Staphylococcus aureus (VISA/hVISA/VRSA) |
gram (+) cocci | skin/soft tissue infections, UTI, bacteremia, toxic shock syndrome, pneumonia, osteomyelitis, endocarditis, meningitis | vancomycin; beta-lactam antibiotics (eg., oxacillin, penicillin, nafcillin, amoxicillin, and most cepholosporins) erythromycin | linezolid; clindamycin; daptomycin; TMP-SMX; quinupristine-dalfopristin |
| Community-acquired methicillin-resistant Staphylococcus aureus (cMRSA) | gram (+) cocci |
necrotizing pneumonia; skin infections, boils, abcesses (seen in IV drug abusers, athletes who share equipment, day care centers, military personnel; prisons); drainage of abscess is primary treatment; treat with antibiotic only if needed | beta-lactam antibiotics (eg., oxacillin, penicillin, amoxicillin, and most cepholosporins, erythromycin | doxycycline or minocycline; clindamycin (confirm with D-test); linezolid; TMP-SMX |
| Streptococcus pneumoniae (multi-drug resistant) | gram (+) diplo- coccus |
pneumonia, otitis media, sinusitis, bronchitis, bacteremia, peritonitis, cellulitis, meningitis, arthritis | multi-drug resistance; penicillin G, cephalosporins, TMP-SMX, erythromycin, doxycycline |
for multi-drug resistance consider: vancomycin +/- rifampin; fluoroquinolone (gemifloxacin, moxifloxacin), levofloxacin)–alternatives: linezolid; clindamycin; imipenem/cilastatin |
| Escherichia coli (E. Coli) – CTX-M extended spectrum beta-lactamases (ESBL) | gram (-) rod |
UTIs | Oral cephalosporins, TMP/SMX, fluoroquinolones | Fosfomycin, nitrofurantoin, ertapenem, doripenem, imipenem/cilastatin |
| Enterococcus faecium (E. faecium) vancomycin resistant enterococci (VRE) |
gram (+) cocci |
meningitis, UTI, bacteremia (central venous catheter-related), endocarditis | vancomycin; streptomycin; gentamicin; penicillin; ampicillin | linezolid; quinupristine-dalfopristin; daptomycin, fosfomycin (for UTI) |
| Pseudomonas aeruginosa (multidrug resistant strains) | gram (-) rod |
UTIs, pneumonias, skin and soft-tissue infections, endocarditis, meningitis | imipenem/cila- statin, mero- penem, non-antipseudo-monal penicillins, oral cephalosporins, |
colistin, polymyxin B (for multidrug resistant strains) |
| Klebsiella pneumoniae -extended spectrum beta-lactamases (ESBL) |
gram (-) rod |
pneumonias, UTIs, upper respiratory tract infections, surgical wound infections | 2nd, 3rd generation cephalosporins; aztreonam; carbapenems | imipenem; meropenem; colistin |
| multi-drug resistant Mycobacterium tuberculosis (MDR-TB) | acid-fast | tuberculosis (lung infection) | isoniazid; rifampin; possibly streptomycin | multiple agents required for treatment: aminoglycoside (amikacin or kanamycin) or polypeptide antibiotic (capreomycin) + antimycobacterials (pyrazinamide + ethambutol) + fluorquinolone (moxifloxacin) + rifabutin; other agents may need to be substituted based on drug availability |
| Acinetobacter baumanii | gram (-) rod |
immunocompromised patients: pneumonia (commonly ventilator-associated), UTI, septicemia, central venous catheter-related infections, traumatic wound infections | imipenem; meropenem; antipseudomonal agents, fluoroquinolones, carbapenems | ampicillin-sulbactam; colistin |
| Staphylococcus epidermidis (methicillin resistant) | gram (+) | bacteremia, catheter, implant, and prostheses-related infection (biofilm formations), endocarditis | penicillin, amoxicillin | vancomycin –if infected implant, surgical removal or replacement may be required; vancomycin +/- (rifampin + gentamicin)–alternative regimens if vancomycin resistant: daptomycin, linezolid |
* Note:
This table is not a comprehensive listing of all resistant bacteria and treatments. Antibiotic resistance patterns are constantly evolving and bacteria may not always exhibit resistance to select antibiotics in every patient. In all cases, antibiotic selection should be based on site of infection and clinical presentation as evaluated by a health care professional, culture/sensitivity and other needed laboratory results, local resistance/susceptibility patterns, and patient-specific characteristics.. In many instances, the care of a team of healthcare providers, including an infectious disease specialist, may be required.
___
___
Resistant bacteria of international concern:
| Escherichia coli | Resistance to third-generation cephalosporins; Resistance to fluoroquinolones |
| Klebsiella pneumoniae | Resistance to third-generation cephalosporins; Resistance to carbapenems |
| Staphylococcus aureus | Resistance to methicillin (MRSA) |
| Streptococcus pneumoniae | Resistance, or non-susceptibility, to penicillin |
| Non-typhoidal salmonella (NTS) | Resistance to fluoroquinolones |
| Shigella species | Resistance to fluoroquinolones |
| Neisseria gonorrhoeae | Decreased susceptibility to third-generation cephalosporins |
| Enterococcus faecalis | Resistance to vancomycin (VRE), to aminopenicillins |
| Pseudomonas aeruginosa | Resistance to carbapenems, to amikacin, to ceftazidime |
| Acinetobacter baumannii | Resistance to carbapenems, to third-generation cephalosporins |
_
_
Staphylococcus aureus:
Staphylococcus aureus (colloquially known as “Staph aureus” or a “Staph infection”) is one of the major resistant pathogens. Found on the mucous membranes and the human skin of around a third of the population, it is extremely adaptable to antibiotic pressure. It was one of the earlier bacteria in which penicillin resistance was found—in 1947, just four years after the drug started being mass-produced. Methicillin was then the antibiotic of choice, but has since been replaced by oxacillin because of significant kidney toxicity. Methicillin-resistant Staphylococcus aureus (MRSA) was first detected in Britain in 1961, and is now “quite common” in hospitals. MRSA was responsible for 37% of fatal cases of sepsis in the UK in 1999, up from 4% in 1991. Half of all S. aureus infections in the US are resistant to penicillin, methicillin, tetracycline and erythromycin. This left vancomycin as the only effective agent available at the time. However, strains with intermediate (4–8 μg/ml) levels of resistance, termed glycopeptide-intermediate Staphylococcus aureus (GISA) or vancomycin-intermediate Staphylococcus aureus (VISA), began appearing in the late 1990s. The first identified case was in Japan in 1996, and strains have since been found in hospitals in England, France and the US. The first documented strain with complete (>16 μg/ml) resistance to vancomycin, termed vancomycin-resistant Staphylococcus aureus (VRSA) appeared in the United States in 2002. However, in 2011, a variant of vancomycin has been tested that binds to the lactate variation and also binds well to the original target, thus reinstating potent antimicrobial activity. A new class of antibiotics, oxazolidinones, became available in the 1990s, and the first commercially available oxazolidinone, linezolid, is comparable to vancomycin in effectiveness against MRSA. Linezolid-resistance in S. aureus was reported in 2001. Community-acquired MRSA (CA-MRSA) has now emerged as an epidemic that is responsible for rapidly progressive, fatal diseases, including necrotizing pneumonia, severe sepsis, and necrotizing fasciitis. Outbreaks of CA-MRSA infections have been reported in correctional facilities, among athletic teams, among military recruits, in newborn nurseries, and among men that have sex with men. CA-MRSA infections now appear endemic in many urban regions and cause most CA-S. aureus infections.
__
Extended-Spectrum beta-lactamase (ESBL) – producing Gram-negative bacteria:
Extended-spectrum beta-lactamases (ESBLs) are plasmid-associated beta lactamases that have recently been found in the Enterobacteriaceae. ESBLs are capable of hydrolyzing penicillins, many narrow spectrum cephalosporins, many extended-spectrum cephalosporins, oxyimino-cephalosporins (cefotaxime, ceftazidime), and monobactams (aztreonam). Beta-lactamase inhibitors (e.g. clavulanic acid) generally inhibit ESBL producing strains. ESBL producing isolates are most commonly Klebsiella ssp, predominantly Klebsiella pneumoniae, and E. coli, but they have been found throughout the Enterobacteriaeae. Because ESBL enzymes are plasmid mediated, the genes encoding these enzymes are easily transferable among different bacteria. Most of these plasmids not only contain DNA encoding ESBL enzymes but also carry genes conferring resistance to several non-ß-Lactam antibiotics. Consequently, most ESBL isolates are resistant to many classes of antibiotics. The most frequent coresistances found in ESBL-producing organisms are aminoglycosides, fluoroquinolones, tetracyclines, chloramphenicol, and sulfamethoxazole-trimethoprim. Treatment of these multiple drug-resistant organisms is a therapeutic challenge. ESBL producing strains have been isolated from abscesses, blood, catheter tips, lung, peritoneal fluid, sputum, and throat cultures. They apparently have a world-wide distribution. Rates of isolation vary greatly worldwide and within geographic areas and are rapidly changing over time. Known risk factors for colonization and/or infection with organisms harboring ESBLs include admission to an intensive care unit, recent surgery, instrumentation, prolonged hospital stay and antibiotic exposure, especially to extended-spectrum beta-lactam antibiotics. Use of extended-spectrum antibiotics exerts a selective pressure for emergence of ESBL producing strains. The resistance plasmids can then be transferred to other bacteria, not necessarily of the same species, conferring resistance to them. The lower GI tract of colonized patients is the main reservoir of these organisms. Gastrointestinal carriage can persist for months. In some cities in the United States, nursing homes may be an important reservoir of ESBL producing strains. Nursing home patients are more likely to be treated empirically with antibiotics, and thus on admission to a hospital to be more likely to possess an ESBL producing strain. Patient to patient transmission of ESBL producing organisms occurs via the hands of hospital staff. It is known that ESBL producing strains can survive in the hospital environment. Nosocomial infections in patients occur through the administration of extended spectrum beta-lactam antibiotics or via transmission from other patients via health care workers, who become colonized with resistant strains via exposure to patients or other health care workers. Spread of ESBL producing strains can be minimized by good infection control practices, especially by good hand washing technique.
_
The Special Problem of Clostridium difficile:
Clostridium difficile is the leading cause of health care–associated infections in the United States and is a cause of epidemics of nosocomial infections. The bacterium’s resistance to multiple antibiotics allows its selection for overgrowth in the gut when the gut microbiome is disrupted by antibacterial drugs. Clostridium difficile spores shed by infected or colonized patients persist on the surfaces of objects in hospitals and may be ingested by patients receiving antibiotics and other therapies. In addition, the hypervirulent strain BI/NAP1/027 possesses increased resistance to fluoroquinolones, giving it a selective advantage in patients treated with that class of antimicrobials. Clostridium difficile bacteria are generally not invasive; however, these organisms elaborate exotoxins (toxins A and B) that cause mucosal damage in the colon leading to the morbidity of the infection. The disease manifests as colitis with diarrhea and classically a colonic pseudomembrane; in a recent multicenter study, 8% of patients with C difficile infection developed severe complications such as toxic megacolon. Clostridium difficile infection is usually treatable with antimicrobials, such as oral metronidazole for mild to moderate infection and oral vancomycin for more severe infection. Recurrent disease occurs in approximately 20% of patients, underscoring the importance of prevention. Newer antimicrobials, such as fidaxomicin, have some efficacy both in treatment of infection and prevention of relapse. Severe or fulminant disease that does not respond to vancomycin or fidaxomicin may be treated surgically. Fecal microbiota transplant (FMT), or transfer of stool from an individual with a healthy fecal microbiome, is an evidence-based and highly effective treatment, with success rates of 81% to 94%. The high efficacy of FMT has made it a standard approach and an often welcome intervention for patients with recurrent relapses or refractory disease. A variety of preventive agents for C difficile are in clinical trials, including vaccines, monoclonal antibodies, therapeutic agents such as nontoxigenic strains (to prevent recurrence), and toxin-binding compounds.
_
Neisseria gonorrhoeae:
Another resistant organism causing increasing concern is Neisseria gonorrhoeae. Gonorrhea is the second most common reportable communicable disease in the United States (after chlamydial disease). Neisseria gonorrhoeae has developed increasing resistance to oral antibiotics (e.g., azithromycin, fluoroquinolones, and the oral cephalosporin cefixime) previously used to treat this infection; in 2014, 37% of N gonorrhoeae isolates in the United States were resistant to at least 1 antibiotic. Although cefixime resistance in the United States has declined in recent years, the slow, inexorable rise in resistance of N gonorrhoeae has resulted in increasingly narrow treatment options and has led the CDC to declare drug-resistant N gonorrhoeae one of the leading microbial threats to public health. Increasing rates of resistance to oral agents have left ceftriaxone as the last remaining reliable treatment for gonorrhea. Resistance to ceftriaxone has been reported, foreshadowing the need for escalation of doses and new drug combinations to overcome resistance. Treatment guidelines reflect these changes as they move toward use of drug combinations. Current recommended first-line therapy for gonorrhea in the United States is ceftriaxone plus azithromycin, even if nucleic acid testing is negative for Chlamydia trachomatis. Recent clinical studies have identified drug combinations that could be used for salvage treatment of nonresponders, such as azithromycin in combination with either gentamicin or gemifloxacin. Although treating on the basis of antimicrobial susceptibility of N gonorrhoeae in individual patients might help preserve the long-term effectiveness of the remaining antimicrobial armamentarium, rapid molecular detection of resistance is not available for this organism. Treatment for N gonorrhoeae must be provided promptly at the point of care to ensure adherence and minimize transmission.
__
The worldwide emergence of plasmid-mediated quinolone resistance:
Fluoroquinolone resistance is emerging in Gram-negative pathogens worldwide. The traditional understanding that quinolone resistance is acquired only through mutation and transmitted only vertically does not entirely account for the relative ease with which resistance develops in exquisitely susceptible organisms, or for the very strong association between resistance to quinolones and to other agents. The recent discovery of plasmid-mediated horizontally transferable genes encoding quinolone resistance might shed light on these phenomena. The Qnr proteins, capable of protecting DNA gyrase from quinolones, have homologues in water-dwelling bacteria, and seem to have been in circulation for some time, having achieved global distribution in a variety of plasmid environments and bacterial genera. AAC(6′)-Ib-cr, a variant aminoglycoside acetyltransferase capable of modifying ciprofloxacin and reducing its activity, seems to have emerged more recently, but might be even more prevalent than the Qnr proteins. Both mechanisms provide low-level quinolone resistance that facilitates the emergence of higher-level resistance in the presence of quinolones at therapeutic levels. Much remains to be understood about these genes, but their insidious promotion of substantial resistance, their horizontal spread, and their co-selection with other resistance elements indicate that a more cautious approach to quinolone use and a reconsideration of clinical breakpoints are needed.
____
Organisms Resistant to Carbapenems:
β-Lactamases are a family of AMR enzymes that hydrolyze β-lactam rings, structures present in common antibiotics such as penicillins, cephalosporins, and aztreonam. Some are considered “extended-spectrum β-lactamases” because they can inactivate a wide range of β-lactam antibiotics. Carbapenemases are even more versatile members of the β-lactamase family due to their ability to hydrolyze both traditional β-lactam antibiotics and carbapenems, the latter representing the broadest-spectrum antibiotics available for treatment of gram-negative bacterial infections. Although many β-lactamase genes are encoded in the bacterial chromosome, extended-spectrum β-lactamase and carbapenemase genes, which render gram-negative bacteria resistant to important antibiotic classes, are usually plasmid mediated. Chromosomally encoded carbapenemases have been recognized for decades; however, only within the past 15 years have plasmid-mediated carbapenemases become clinically significant. Enteric bacteria carrying the K pneumoniae carbapenemase gene (blaKPC) were first reported in the United States in the early 2000s. They became widespread in health care facilities in the northeastern states and then in Israel. Within a decade, blaKPC variants and several additional plasmid-carried carbapenemases were identified in other regions of the world, typically in health care–associated gram-negative bacteria. It soon became apparent that infections with these multidrug-resistant organisms were associated with mortality rates of 40% to 80%.
NDM-1
The resistance is bestowed by a gene, blaNDM-1, that encodes the enzyme New Delhi metallo-β-lactamase 1 (NDM-1). These genes can be passed easily between bacteria by discrete rings of DNA called plasmids. The enzyme blocks the activity of a range of antibiotics including the carbapenems — drugs of last resort for resistant infections — which might be used to treat, for example, urinary-tract infections triggered by the bacterium Escherichia coli or lung infections resulting from Klebsiella pneumoniae. The New Delhi metallo-β-lactamase (NDM-1)–containing bacteria were identified in India in 2008 and rapidly became endemic throughout South Asia and the Balkan states. The NDM-1 gene has now been implicated in nosocomial infections and outbreaks on every inhabited continent. Bacteria containing NDM-1 gene are so widespread on the Indian subcontinent that they have been cultured from runoff water on the streets, in newborns delivered in hospitals, and in community-acquired infections. The NDM-1 gene has been identified in a broad range of gram-negative bacteria beyond enteric flora, including Acinetobacter and Pseudomonas. The gene for NDM-1 is one member of a large gene family that encodes beta-lactamase enzymes called carbapenemases. Bacteria that produce carbapenemases are often referred to in the news media as “superbugs” because infections caused by them are difficult to treat. The most common bacteria that make this enzyme are Gram-negative such as Escherichia coli and Klebsiella pneumoniae, but the gene for NDM-1 can spread from one strain of bacteria to another by horizontal gene transfer. The NDM-1 enzyme was named after New Delhi, the capital city of India, as it was first described by Yong et al. in December 2008 in a Swedish national who fell ill with an antibiotic-resistant bacterial infection that he acquired in India. The infection was identified as a carbapenem-resistant Klebsiella pneumoniae strain bearing the novel gene NDM-1. The authors concluded that the new resistance mechanism clearly arose in India, but there are few data arising from India to suggest how widespread it is. Its exact geographical origin, however, has not been conclusively verified. The Indian health ministry has disputed the conclusion of Lancet study that the gene originated in India, describing this conclusion as “unfair” and stating that Indian hospitals are perfectly safe for treatment. However, Deshpande P and team from Hinduja National hospital, Mumbai have isolated 22 NDM-1 producing Enterobacteriaceae, from span of just 3 months and a single hospital. If a single hospital can isolate such a significant number of bacteria with a new resistance gene in a short period of time; the data from all the Indian hospitals, if available would be shocking. Enterobacteriaceae with NDM-1 carbapenemases are highly resistant to many antibiotic classes and potentially herald the end of treatment with β-lactams, fluoroquinolones, and aminoglycosides—the main antibiotic classes for the treatment of Gram-negative infections. Many NDM-1 strains are resistant to all antibiotics except for colistin. Colistin is an older antibiotic that has not been used much in recent decades, because it is somewhat more toxic than other antibiotics. A few NDM-1 strains have been sensitive to tigecycline, but this agent should be used cautiously in serious infections because it does not achieve high levels in the bloodstream. A few strains have also been sensitive to aztreonam. The spread of NDM-1 within health-care facilities can be curbed through strict infection-control measures, including patient isolation and hand washing.
_
Organisms Resistant to Colistin:
MCR-1
MCR-1 is a genetic mechanism by which the mcr-1 gene confers the first known plasmid-mediated resistance to colistin, a polymixin and one of a number of last-resort antibiotics. The mechanism, first discovered in E. coli (strain SHP45) from a pig in China in November 2015, was later found by independent researchers in human samples from Malaysia, England, China, Europe and the United States. MCR-1 is the first known polymixin resistance mechanism capable of horizontal gene transfer. On 26 April 2016, a 49-year-old woman sought medical care at a Pennsylvania clinic for UTI symptoms. PCR of an E. coli isolate cultured from her urine revealed the first known presence of the mcr-1 gene in the United States. These findings emphasise the urgent need for coordinated global action in the fight against pan-drug-resistant Gram-negative bacteria. Since its discovery, mcr-1 has been identified in Enterobacteriaceae cultured from humans, animals, and meat in at least 5 continents, including North America. As evidenced by an isolate reported from Germany, when this gene finds its way to a highly resistant carbapenemase-producing organism, the result would be a pan-resistant organism that is potentially untreatable with any existing antimicrobial drugs. Recently Indian researchers have isolated a strain of E.Coli bacteria, carrying MCR -1 gene, described previously as ‘truly pan-drug resistant’. It is resistant to the last mile antibiotic the human race currently has access to —colistin. While colistin resistance had already been detected in India, it existed thus far only as mutations in the genetic path. With MCR-1, however, the gene is found in the plasmid medium, a small DNA molecule outside of the chromosomal DNA, meaning the resistance can spread in hospitals, and the community.
__________
Resistant Fungi:
Infections by fungi are a cause of high morbidity and mortality in immunocompromised persons, such as those with HIV/AIDS, tuberculosis or receiving chemotherapy. The fungi candida, Cryptococcus neoformans and Aspergillus fumigatus cause most of these infections and antifungal resistance occurs in all of them. Multidrug resistance in fungi is increasing because of the widespread use of antifungal drugs to treat infections in immunocompromised individuals. Increased prevalence and duration of fungal infections in many clinical settings, together with the refractory nature of some fungi to treatment, have made fungal susceptibility testing a necessary component of the microbiology laboratory. Amphotericin B, the first agent to be discovered, is considered the most effective antifungal for invasive infections involving mold or yeast. It acts by binding to ergosterol in the fungal cell membranes, resulting in the formation of holes in the membranes, to an osmsotic imbalance, and, in the end, to lysis of the cell. Resistance to amphotericin B is considered rare. In fact, whether true resistance exists or whether an occassional lack of efficacy is related to host or environmental factors remains a topic of debate. The imidazoles, introduced in the 1960s, block the synthesis of ergosterol, thereby disrupting the fungal cell membrane structure and inhibiting growth of the organism. They include ketoconazole and miconazole, the latter of which is seldom used today because it must be administered intravenously and is fairly toxic. Ketoconazole has a wide spectrum of activity against molds and filamentous fungi. The triazoles (ie, fiuconazole and itraconazole), licensed for general use in the 1990s, have the same mode of action as that of the imidazoles. Fluconazole is water soluble and has good CSF penetration. This drug is used largely to treat Candida spp and cryptococcal meningitis in people with AIDS. Resistance has been reported in some yeasts including Candida albicans, Candida krusei, and Candida glabrata. A variety of mechanisms can lead to acquired resistance of Candida species to azole drugs, the most common being induction of the efflux pumps encoded by the MDR or CDR genes, and acquisition of point mutations in the gene encoding for the target enzyme (ERG11). Acquired resistance of Candida species to echinocandins is typically mediated via acquisition of point mutations in the FKS genes encoding the major subunit of its target enzyme. Antifungal resistance is associated with elevated minimum inhibitory concentrations, poorer clinical outcomes, and breakthrough infections during antifungal treatment and prophylaxis. Susceptibility testing methods for fungi versus antifungal agents are almost identical to those used for bacterial testing. They include micro, macro, and agar dilution, as well as the E-test.
_____
Resistant Viruses:
Specific antiviral drugs are used to treat some viral infections. These drugs prevent viruses from reproducing by inhibiting essential stages of the virus’s replication cycle in infected cells. Antivirals are used to treat HIV, hepatitis B, hepatitis C, influenza, herpes viruses including varicella zoster virus, cytomegalovirus and Epstein-Barr virus. With each virus, some strains have become resistant to the administered drugs. There are two antiviral drugs used to treat seasonal influenza; oseltamivir and zanamivir, with the recommended treatment being dependent upon the circulating stain. Treatment prevents serious infection and lessens the duration of symptomatic illness. Both are neuraminidase (NA) inhibitors, binding to the virus’s NA surface protein and inhibiting enzymatic activity. This prevents flu viruses from spreading from infected cells to other healthy cells. Resistance can emerge through one of numerous mutations in the NA that reduce inhibitor binding efficiency to the enzyme. These mutations differ between type and subtype of influenza virus and in their impact on the two inhibitors, but the most prevalent is an H275Y mutation, known to confer oseltamivir resistance in 2009 H1N1 flu viruses. Treatment of chronic hepatitis C virus (HCV) infections has changed dramatically since the introduction of direct acting antivirals (DAA). The treatments telaprevir and boceprevir have been supplemented by new NS3 protease inhibitors, simeprevir and faldaprevir, a non-nucleoside polymerase inhibitor, sofosbuvir and NS5a replication complex inhibitors daclatasvir and ledipasvir. Used in combination, these have greatly increased the treatment options for chronic HCV with high efficacy and improved safety. Naturally occurring mutations such as the Q80K variant conferring resistance to simeprevir has been observed in proportions ranging from 9%-48% of untreated HCV genotype 1a-infected patients. Resistant variants are detectable in the majority of patients with treatment failure to NS3 protease inhibitor- or NS5a inhibitor-based antiviral therapy. Long-term follow-up studies by population-based sequence analysis have shown the disappearance of resistant variants in the majority of affected patients, with median times to the disappearance of these types of mutations of 4-64 weeks. Resistance to HIV antivirals is discussed below.
______
______
Overview of AMR to tuberculosis (TB), HIV and malaria therapy:
_
_
Resistant tuberculosis:
AMR in TB vs. other bacteria:
The need to stem the growing problem of antimicrobial resistance has prompted multiple, sometimes conflicting, calls for changes in the use of antimicrobial agents. One source of disagreement concerns the major mechanisms by which antibiotics select resistant strains. For infections like tuberculosis, in which resistance can emerge in treated hosts through mutation, prevention of antimicrobial resistance in individual hosts is a primary method of preventing the spread of resistant organisms in the community. By contrast, for many other important resistant pathogens, such as penicillin-resistant Streptococcus pneumoniae, methicillin-resistant Staphylococcus aureus, and vancomycin-resistant Enterococcus faecium, resistance is mediated by the acquisition of genes or gene fragments by horizontal transfer; resistance in the treated host is a relatively rare event. For these organisms, indirect, population-level mechanisms of selection account for the increase in the prevalence of resistance. These mechanisms can operate even when treatment has a modest, or even negative, effect on an individual host’s colonization with resistant organisms. For many pathogens of current concern, especially organisms for which asymptomatic colonization typically precedes infection (e.g., Streptococcus pneumoniae, Staphylococcus aureus, Enterococcus spp., and the gram-negative enteric bacteria), the relationship between antimicrobial use and resistance differs in fundamental ways from the relationship found in Mycobacterium tuberculosis, for which many modern principles of chemotherapy were developed. Ehrlich’s advice that treatment of infections should “hit hard and hit early,” formulated in the earliest days of antimicrobial chemotherapy, presciently summarized the principles of treatment for infections such as tuberculosis (TB). These principles are embodied in modern protocols of directly observed, short-course chemotherapy, where the goal is to treat with adequate concentrations of multiple drugs and maintain treatment until the bacterial population is extinct. Resistance to each of the major antituberculosis drugs is mediated by single point mutation; therefore tuberculosis treatment is designed to prevent the ascent of subpopulations of mutant bacilli that are resistant to any one of the drugs. Similar principles have been suggested for other infections in which resistance can arise by simple mutation, most notably HIV, although there has been some controversy on this topic. In these infections, the relationship between treatment, resistance in the treated person, and resistance in the community at large is relatively clear. Inadequate therapy (owing to subtherapeutic drug concentrations, too few drugs, or poor adherence to therapy) results in the emergence of resistance, and possibly treatment failure, in the treated host. Following the emergence of resistance in the treated host, resistant infections may be transmitted to others.
_
Types of Drug-Resistant TB:
1. Multidrug-Resistant TB (MDR TB)
Multidrug-resistant TB (MDR TB) is caused by TB bacteria that is resistant to at least isoniazid and rifampin, the two most potent TB drugs. These drugs are used to treat all persons with TB disease.
2. Extensively Drug-resistant TB (XDR TB)
Extensively drug-resistant TB (XDR TB) is a rare type of MDR TB that is resistant to isoniazid and rifampin, plus any fluoroquinolone and at least one of three injectable second-line drugs (i.e., amikacin, kanamycin, or capreomycin). Because XDR TB is resistant to the most potent TB drugs, patients are left with treatment options that are much less effective. XDR TB is of special concern for people with HIV infection or other conditions that can weaken the immune system. These people are more likely to develop TB disease once they are infected, and also have a higher risk of death once they develop TB.
3. Totally drug resistant TB (TDR TB)
TB is resistant to all drugs of first and second line treatment. Dr. Zarir Udwadia, whose group from the P D Hinduja National Hospital and Medical Research Centre, Mumbai, was the first to describe “totally drug resistant TB” in patients in India in 2011, says that India had made the transition from a place of almost non-existent treatment of MDR-TB to one where these patients could now be treated. However, his concern was that “the numbers enrolled remain minuscule compared with the numbers crying out for effective treatment, and the programme needs to expand rapidly so more patients are included.” Dr. Udwadia hoped that the widespread use of a new diagnostic test for TB called the GeneXpert MTB/RIF would help benefit patients, as it drastically cuts time to diagnosis and can also detect rifampicin resistance.
_
The figure below shows that India and China have highest number of MDR TB:
_
In 2014 the World Health Organisation (WHO) estimated that 3.3% of new cases and 20% of previously treated cases of TB were of MDR TB. Among patients with pulmonary TB who were notified to WHO in 2014 it is estimated that 300,000 had MDR-TB. Globally, only half of MDR-TB patients were successfully treated in 2014. More than half of these patients were in India, China and the Russian Federation. Extensively drug-resistant tuberculosis (XDR-TB), a form of tuberculosis that is resistant to at least 4 of the core anti-TB drugs, has been identified in 105 countries. An estimated 9.7% of people with MDR-TB have XDR-TB. Some organisations believe that the current statistics for drug resistant TB greatly underestimate the extent of the problem. Traditionally the view in India has been that drug resistant TB is not easily transmissible. So it is believed that most drug resistant TB in India arises from the failure of people to take their drugs properly, rather than from them becoming infected with an MDR TB strain. Treatment of drug resistant TB is further complicated by the decreased efficacy and higher toxicity associated with the second line drugs as well as the inability to provide early diagnostic data to guide treatment. Conventional drug susceptibility testing relies on mycobacterial culture methods, providing results after weeks or months. Despite remarkable strides made in advancing diagnosis of drug-resistant TB with molecular-based diagnostics, such as the GeneXpert MTB/RIF assay, this technology currently detects resistance to RIF only.
_
Drug resistance is not acquired through horizontal gene transfer in M. tuberculosis, since this pathogen does not contain plasmids and the transfer of genomic DNA has not been demonstrated. Thus, resistance to anti-TB drugs develops by spontaneous mutation and the resulting resistant mutants are selected by subsequent treatment with anti-TB drugs to which the mutants are resistant.
_
Causes of Drug-Resistant TB:
Drug-resistant TB can occur when the drugs used to treat TB are misused or mismanaged.
Examples of misuse or mismanagement include
•People do not complete a full course of TB treatment
•Health care providers prescribe the wrong treatment (the wrong dose or length of time)
•Drugs for proper treatment are not available
•Drugs are of poor quality
_
Drug-resistant TB is more common in people who
•Do not take their TB drugs regularly
•Do not take all of their TB drugs
•Develop TB disease again, after being treated for TB disease in the past
•Come from areas of the world where drug-resistant TB is common
•Have spent time with someone known to have drug-resistant TB disease
_
Best drug treatment for multidrug-resistant and extensively drug-resistant tuberculosis:
Multidrug-resistant (MDR) and extensively drug-resistant (XDR) tuberculosis are generally thought to have high mortality rates. However, many cases can be treated with the right combination and rational use of available antituberculosis drugs. The recommended regimen is the combination of at least four drugs to which the Mycobacterium tuberculosis isolate is likely to be susceptible. Drugs are chosen with a stepwise selection process through five groups on the basis of efficacy, safety, and cost. Among the first group (the oral first-line drugs) high-dose isoniazid, pyrazinamide, and ethambutol are thought of as an adjunct for the treatment of MDR and XDR tuberculosis. The second group is the fluoroquinolones, of which the first choice is high-dose levofloxacin. The third group are the injectable drugs, which should be used in the following order: capreomycin, kanamycin, and amikacin. The fourth group are called the second-line drugs and should be used in the following order: thioamides, cycloserine, and aminosalicylic acid. The fifth group includes drugs that are not very effective or for which there are sparse clinical data. Drugs in group five should be used in the following order: clofazimine, amoxicillin with clavulanate, linezolid, carbapenems, thioacetazone, and clarithromycin.
_
New Drug available to treat Multidrug-Resistant Tuberculosis:
Bedaquiline is the first drug in a new class of anti-TB medications to be approved in more than 40 years by the US Food and Drug Administration (FDA). It is important to note, however, that owing to the potential for severe adverse events, bedaquiline is not recommended for all patients with MDR TB. Bedaquiline may be used for 24 weeks of treatment in adults with laboratory-confirmed pulmonary MDR TB when an effective treatment regimen cannot otherwise be provided. The current recommended dose of bedaquiline is 400 mg given orally with food, by directly observed therapy, once daily for 2 weeks; followed by 200 mg given 3 times per week for an additional 22 weeks. To lessen the chance for TB bacteria to become resistant to bedaquiline, this antibiotic should only be used in combination with at least 3-4 other antibiotics to which laboratory tests indicate that the bacteria is susceptible. Bedaquiline may be used on a case-by-case basis in children, HIV-infected persons, pregnant women, persons with extrapulmonary MDR TB, and patients with comorbid conditions on medications, when an effective treatment regimen cannot otherwise be provided. Patients who take bedaquiline should be monitored closely for suspected and severe adverse events.
__
Detection and treatment of multidrug resistant TB in India remains low: BMJ 2013:
Only 6% of the estimated 66 000 cases of multidrug resistant tuberculosis (MDR-TB) in India in 2011 were detected, and only 5% of these patients were enrolled in second line treatment programmes, a study has found. This study said that a number of factors contributed to poor detection, including lack of awareness and poor access to drug susceptibility testing; inadequate data management of laboratory test results; inexperience of healthcare workers; and a broad spectrum of resistance among patients. Difficulties and delays in obtaining and delivering second line drugs and poor compliance with the treatment regimen also contributed to poor results.
______
______
Resistant malaria:
Development of Resistance:
The malaria parasite is well known for its frequent, de novo mutations, mostly single, and sometimes multiple. In the presence of heavy infection and inadequate drug levels, the resistant mutations survive and propagate. Development of resistance requires a high grade of parasitemia, coupled with low or inadequate drug levels. Most cases of resistance have emerged out of SE Asia region. This region is known for low transmission and low immunity that lead to high parasitemia; it also has a long history of indiscriminate use of different antimalarial drugs. In such low-transmission areas, most malaria infections are symptomatic, and therefore proportionally more people receive treatment, providing more opportunities for selection of resistant strains. One study also suggested that P. falciparum in South-East Asia has an inherent propensity to develop drug resistance through genetic mutation. Areas with very high transmission, such as Africa, appear to be less susceptible to the emergence of drug resistance. In these areas, infections are acquired repeatedly throughout life, resulting in partial immunity, that in turn controls the infection, usually at levels below those that cause symptoms. Asymptomatic infection and often non-availability of drugs in these areas mean that these patients do not receive antimalarial drugs, and hence the chances of development of resistance are lower. Immunity acts by non-selectively eliminating blood-stage parasites, including the rare de novo resistant mutants, and also improves cure rates, even with failing drugs, thereby reducing the relative transmission advantage of resistant parasites. Furthermore, complex polyclonal infections in semi-immune people allow possible outbreeding of multigenic resistance mechanisms or competition in the host or the mosquito between less-fit resistant strains and more-fit sensitive strains. Drug pressure leads to higher gametocyte release, and this facilitates the propagation of the resistant mutants that have escaped the drugs. Failure to use primaquine as a gametocytocidal agent for P. falciparum further aids such spread of resistance. Therefore, gametocyte production from the recrudescent resistant infection must be prevented by administration of early, appropriate treatment, combined with primaquine. Administration of drugs with long elimination phases facilitates the spread of resistant mutant malaria parasites. The residual antimalarial activity that is present during the post-treatment period serves as a “selective filter”, which prevents infection by sensitive parasites but allows infection by resistant parasites. Drugs such as chloroquine, mefloquine and piperaquine, which persist in the blood for months, provide a selective filter long after their administration has ceased. Factors that promote the development of drug resistance are more intense with P. falciparum compared to P. vivax and this explains the higher incidence of resistance in P. falciparum.
_
As of July 2016, resistance to the first-line treatment for P. falciparum malaria (artemisinin-based combination therapies, also known as ACTs) has been confirmed in 5 countries of the Greater Mekong subregion (Cambodia, the Lao People’s Democratic Republic, Myanmar, Thailand and Viet Nam). In most places, patients with artemisinin-resistant infections recover fully after treatment, provided that they are treated with an ACT containing an effective partner drug. However, along the Cambodia-Thailand border, P. falciparum has become resistant to almost all available antimalarial medicines, making treatment more challenging and requiring close monitoring. There is a real risk that multidrug resistance will soon emerge in other parts of the subregion as well. The spread of resistant strains to other parts of the world could pose a major public health challenge and jeopardize important recent gains in malaria control. Artemisinin resistance has occurred as a consequence of several factors: poor treatment practices, inadequate patient adherence to prescribed antimalarial regimens, and the widespread availability of oral artemisinin-based monotherapies and substandard forms of the drug. The geographic scope of the problem could widen quickly and have dire public health consequences: the spread or independent emergence of artemisinin resistance in other parts of the world could pose a major health security risk as no alternative antimalarial medicine is available at present with the same level of efficacy and tolerability as ACTs.
_
Timeline of antimalarial resistance:
| Drug | Introduction | “First” year resistance reported | Difference (years) |
| Quinine | 1632 | 1910 | 278 |
| Chloroquine | 1945 | 1957 | 12 |
| Proguanil | 1948 | 1949 | 1 |
| Sulfadoxine-pyrimethamine | 1967 | 1967 | <1 |
| Mefloquine | 1977 | 1982 | 5 |
| Atovaquone | 1996 | 1996 | <1 |
| Artemisinins | 1971 | 1998? | 35 |
_
Mechanisms of resistance:
Resistance is mediated by transporter mutations. The P. falciparum genome encodes multiple predicted transporters. The biochemical mechanism of resistance has been well understood in cases of chloroquine, the antifolates, and atovaquone. The chloroquine-resistant strains of P. falciparum tend to accumulate the drug less efficiently than the sensitive ones. Polymorphism in the pfcrt (for chloroquine resistance transporter) gene, particularly the one amino acid change, K76T, located in the first transmembrane domain, has been found consistently in chloroquine-resistant P. falciparum parasites. This critical K76T mutation could possibly alter the selectivity of CRT such that chloroquine more efficiently exits the food vacuole. Another mutation could be at the pfmdr1 gene encoding for the transporter for importing solutes into the food vacuole, including the drugs mefloquine, halofantrine, and artemisinin (and possibly chloroquine), but this may not confer resistance on its own. Resistance to quinine, the oldest antimalarial drug, was reported first in Brazil and later in southeast Asia. Quinine resistance is associated with polymorphisms in several transporters. In the laboratory, P. falciparum resistance to chloroquine can be reversed by combining it with various drugs such as calcium inhibitors, phenothiazines, antidepressants, and antihistamine compounds, but clinical evidence is limited and the usefulness of this approach in humans has not been established.
_______
_______
Resistant HIV:
In 2010, an estimated 7% of people starting antiretroviral therapy (ART) in developing countries had drug-resistant HIV. In developed countries, the same figure was 10–20%. Some countries have recently reported levels at or above 15% amongst those starting HIV treatment, and up to 40% among people re-starting treatment. This requires urgent attention. Increasing levels of resistance have important economic implications as second and third-line regimens are 3 times and 18 times more expensive, respectively, than first-line drugs. Since September 2015, WHO has recommended that everyone living with HIV start on antiretroviral treatment. Greater use of ART is expected to further increase ART resistance in all regions of the world. To maximize the long-term effectiveness of first-line ART regimens, and to ensure that people are taking the most effective regimen, it is essential to continue monitoring resistance and to minimize its further emergence and spread. In consultation with countries, partners and stakeholders, WHO is currently developing a new “Global Action Plan for HIV Drug Resistance (2017-2021)”.
_
Antiviral drug resistance is defined by the presence of viral mutations that reduce drug susceptibility compared with the susceptibility of wild-type viruses. Antiviral resistance can be mediated either by changes in the molecular target of therapy (the primary mechanism observed in HIV-1) or in other viral proteins that indirectly interfere with a drug’s activity. HIV-1 drug resistance should be distinguished from other causes of drug failure such as nonadherence, insufficient drug levels, and drug regimens with intrinsically weak antiviral activity. The terms “drug resistance” and “reduced drug susceptibility” have similar meanings, provided that each term is viewed as representing a continuum between susceptible and highly resistant. Because antiretroviral drugs differ in their potencies, reductions in susceptibility must be related to the activity of the drug against wild-type viruses. Pre-existing resistant variants are often present in a small subset of wild-type virus populations. Although many group O isolates are intrinsically resistant to NNRTIs, naturally occurring resistance in group M HIV-1 is uncommon for currently approved antiretroviral drugs
_
The use of combinations of antiretroviral drugs has proven remarkably effective in controlling the progression of human immunodeficiency virus (HIV) disease and prolonging survival, but these benefits can be compromised by the development of drug resistance. Resistance is the consequence of mutations that emerge in the viral proteins targeted by antiretroviral agents. In the United States, as many as 50 percent of patients receiving antiretroviral therapy are infected with viruses that express resistance to at least one of the available antiretroviral drugs. Consequently, the transmission of drug-resistant strains is also a growing concern. Because drug-resistant HIV often exhibits resistance to several classes of antiretroviral drugs and because cross-resistance between drugs within a class is frequent, the emergence of resistance always complicates further efforts to control viral replication.
_
Mechanisms of HIV resistance:
_
The HIV virus is highly adaptive. This means it changes itself to survive better within its host. HIV replicates very quickly— so quickly it often makes mistakes when it copies itself. Some of these mistakes, or mutations, are beneficial to the virus. Through natural selection, the virus starts purposely including these beneficial mutations when it copies itself. This ultimately results in a new strain—or variation—of the virus. The new strain can make the HIV virus less reactive to the medications you’re taking, which is known as drug resistance. HIV drug resistance can be difficult to treat, so make sure you’re informed about this serious problem. HIV treatment is very different than it was in the past. Now, with the right treatment, people who are HIV-positive live long, healthy lives.
_
HIV experts share their tips for treating your HIV.
1. Drug resistance results when HIV mutates itself.
In the case of drug resistance, the virus can keep replicating despite the presence of the drug. The strains of the virus without the mutation tend to die off, while the strain with the mutation thrives. Once drug resistance develops, it isn’t reversible and it can cause your treatment to fail.
2. Poor medication adherence is the main cause of drug resistance.
Not taking every medicine correctly every day—or poor medication adherence—is the main cause of drug resistance. HIV drugs slow the replication of the virus, but they can’t stop it completely. Skipping a dose gives the HIV virus the chance to multiply faster and possibly develop a drug-resistant mutation. That’s why it’s so important to be ready for the commitment of HIV treatment when you start therapy.
3. You can transmit drug-resistant HIV to someone else.
Once you have a drug-resistant strain of HIV in your blood, it’s possible to transmit it to someone else. It’s also possible for you to get a drug-resistant virus from someone else. In either case, this is called transmitted drug resistance. If a person gets a drug-resistant strain, treating it with that specific drug will not work. So it’s important to know what type of virus you have—drug resistant or “wild type.” A wild type has no drug-resistant mutations.
4. Your doctor can test for drug resistance.
As soon as you know you have HIV, your doctor should order drug-resistance testing. It’s a blood test that can show if the virus has transmitted drug resistance. This will help your doctor choose your first HIV regimen. You should have drug-resistance testing even if you plan to delay HIV treatment. Your doctor should repeat the testing whenever you decide it’s time to start treatment. Your doctor may also order testing if it looks like your HIV regimen is failing. It will help with selecting a new regimen.
5. Drug resistance can apply to an entire class of HIV medicines.
When HIV develops resistance to one drug in a class of HIV medicines, it is sometimes resistant to other drugs in the same class. This is called cross-resistance. It can mean some HIV drugs won’t work for you even if you’ve never taken them. Cross-resistance can limit your drug choices for future HIV regimens.
6. You can reduce the risk of drug resistance.
The good news is you can take steps to decrease the risk of drug resistance. The single most important thing you can do is to take your HIV medicines exactly as directed every single day. HIV regimens involve multiple drugs with different schedules, storage requirements, and food restrictions. This requires a high level of commitment.
________
AMR and immunocompromised:
When you have any infection, your immune system is activated. Often it takes care of infection and prevents disease, for example, latent tuberculosis. If you have pneumonia, the infection itself stimulates the body’s immune system but immunity does not always deal with an infection by itself, especially with serious infections. Antibiotics, when properly prescribed, will kill bacteria, render them unable to multiply, or make them more susceptible to the body’s natural immune defenses. Antibiotics are nothing but synergistic allies with the body’s immune system. Antibiotics work in synergy with immune system to kill microbes. Therefore impairment of immune system makes it harder for antibiotic to kill microbe resulting in AMR. In other words, when all other factors are same, immunocompromised individual is more likely to develop AMR than immunocompetent individual. Additionally besides susceptible to routine pathogens, immunocompromised is highly susceptible to opportunistic pathogens which are likely to be intrinsically resistant to antibiotics. AIDS epidemic has greatly enlarged the population of immunocompromised patients at risk of opportunistic infections, and the resurgence of old foes such as malaria and tuberculosis, causing millions of infections each year. Immunocompromised get frequent infections and frequent antibiotic use along with need for frequent empirical use of antibiotics is associated with antibiotic resistance. Frequent antibiotic use lead to development of resistance gene in commensals and reduction in colonisation resistance. Also, killing commensal bacteria with antibiotics might well weaken the immune system. An example is Staphylococcus epidermidis, a skin commensal. It secretes compounds that activate toll-like receptors in keratinocytes (skin cells) which in turn stimulates the release of antimicrobial peptides. These peptides, which are part of the innate immune system, suppress the growth of potentially pathogenic bacteria. So an antibiotic which wiped out S epidermidis could indeed have negative effects on innate immunity. Another example: bacteria in the gut release polysaccharides which modulate the activity of CD4+ and CD8+ immune cells. In both of these examples commensal bacteria are manipulating the immune response to suppress competitors. If these competitors are also human pathogens, then the interaction is mutually beneficial.
________
________
Causes of antimicrobial resistance:
_
Natural occurrence:
Naturally occurring antibiotic resistance is common. Genes for resistance to antibiotics, like antibiotics themselves, are ancient. The genes that confer resistance are known as the environmental resistome. These genes may be transferred from non-disease-causing bacteria to those that do cause disease, leading to clinically significant antibiotic resistance. In 1952 it was shown that penicillin-resistant bacteria existed before penicillin treatment; and also preexistent bacterial resistance to streptomycin. In 1962, the presence of penicillinase was detected in dormant endospores of Bacillus licheniformis, revived from dried soil on the roots of plants, preserved since 1689 in the British Museum. Six strains of Clostridium, found in the bowels of William Braine and John Hartnell (members of the Franklin Expedition) showed resistance to cefoxitin and clindamycin. Penicillinase may have emerged as a defense mechanism for bacteria in their habitats, such as the case of penicillinase-rich Staphylococcus aureus, living with penicillin-producing Trichophyton; however, this may be circumstantial. Search for a penicillinase ancestor has focused on the class of proteins that must be a priori capable of specific combination with penicillin. The resistance to cefoxitin and clindamycin in turn was attributed to Braine’s and Hartnell’s contact with microorganisms that naturally produce them or random mutation in the chromosomes of Clostridium strains. There is evidence that heavy metals and other pollutants may select for antibiotic-resistant bacteria, generating a constant source of them in small numbers.
_
To adequately address the threat posed by AMR, it is important to understand the factors driving its emergence. For example, bacterial replication cycles enable emergence of de novo mutations: a single Staphylococcus aureus bacterium can replicate through 10 generations in less than 12 hours, producing 1 million progeny. Each replication cycle offers the opportunity for mutation, allowing the emergence of genetic factors that contribute to resistance to antibiotics. Although de novo mutations can cause new problems today, naturally occurring resistance factors appear to predate the antibiotic era. Permafrost samples from the Yukon revealed the presence of bacteria with resistance mutations 30,000 years before the discovery of penicillin. Resistance factors also were identified in samples drawn from a cave ecosystem that was isolated for more than 4 million years. Moreover, phylogenetic analyses of β-lactamases (enzymes that render penicillin like antibiotics ineffective) indicate their emergence 1 billion to 2 billion years ago.
_
Antimicrobial resistance can be caused by “selection pressure.” Regardless of how effective an antimicrobial might be, rarely, if ever, will 100% of the organisms be killed during a course of treatment. This means that at least one organism out of thousands may have developed resistance to the antimicrobial. The few surviving and potentially resistant organisms could then transfer their genetic material to offspring or even other unrelated organisms. There are also some who say that antimicrobial resistance is caused by widespread use of antimicrobials in food production systems. Their argument is that the more antimicrobials are used in animals, the more we expose the organisms to the antimicrobials and give them the opportunity to develop resistance. Although that may be true in a very simplified, general sense, the scientific evidence of how, if or to what extent such exposure affects human health remains unclear.
_
The figure below shows drivers for antimicrobial resistance:
_
Although naturally occurring resistance factors contribute to AMR, antibiotic use selects for their emergence; therefore, human activity plays an important role in the evolution of AMR. For example, consider the agricultural use of antibiotics for promotion of animal growth. In the United States, antibiotic use in animals raised for food represents 80% of total antibiotic consumption. The US Food and Drug Administration (FDA) estimates that 74% of these antibiotics are administered in feed, a method commonly used to promote animal growth, rather than to treat or prevent infection. Moreover, 62% of the antibiotics used in animals represent “medically important” compounds, i.e., they have a role in treating human disease. Antibiotics used in the remaining 38% may influence human health as well. For example, bacitracin is commonly used topically on humans but not systemically administered as it is in animals. Although the direct influence of such practices on human health is difficult to quantify, reports of transmission of resistant bacteria via animal-to-human contact and consumption of animal products continue to emerge. Furthermore, an association has been demonstrated between antibiotic consumption by animals and the existence in humans of commensal organisms resistant to the same antibiotic classes. There also are reports of resistant pathogens passing from humans to animals. The assumption that simply giving antimicrobials to a larger number of animals creates a public health hazard due to resistance isn’t accurate, because it doesn’t account for the benefits of preventing disease and the need for higher doses and potentially stronger types of antimicrobials if an animal is sick. For example, if a disease is not prevented or effectively treated by a low dose of an antimicrobial, a higher dose or a different antimicrobial treatment, or both, are often needed to eliminate the infection. This would obviously increase the amount of antimicrobials used, and it would effectively kill most of the target organisms. Yet, it could potentially increase the development of resistance to a stronger drug in the organism you’re targeting. As previously stated, no antimicrobial is 100% effective and therefore the remaining few bacteria may be resistant and transfer that resistance. The greater dose could also decrease the potential for the development of resistance by minimizing the numbers of remaining bacteria. Yet, you can’t pick and choose which organisms will be exposed to the antimicrobial – organisms that aren’t the targets of that antimicrobial will also be exposed to these high drug doses and stronger drugs. These “innocent bystander” organisms could potentially develop resistance mechanisms and pass on that resistance to their offspring and to other organisms. Sometimes a lower dose is enough to prevent an infection, and this may be the best way to prevent the disease before the entire herd or flock is affected and higher doses or stronger types of antimicrobials must be used. For some, this would translate into “less use” because it’s a lower dose of antimicrobials used, but others would interpret this to mean “more use” because the antimicrobial is given to a larger number of animals.
_
Human antibiotic use also contributes to the emergence of AMR. Rising drug resistance can be attributed to three causes use of antibiotics: in the human population; in the animal population; and spread of resistant strains between human or non-human sources. Antibiotics increase selective pressure in bacterial populations, causing vulnerable bacteria to die—this increases the percentage of resistant bacteria which continue growing. Inpatient antibiotic usage represents only 38.5% of the total antibiotic market. A recent analysis revealed that 12.6% of outpatient visits in the United States resulted in the prescribing of an antibiotic, and 30% of those prescriptions may have been inappropriate. Direct-to-consumer sales compound the problem of inappropriate use in many areas of the world. Outside of the United States and Europe, such purchases account for one-fifth to nearly all antibiotic use, depending on location. Worldwide, antibiotics dispensed directly to the consumer are more likely to be inappropriately selected, taken at doses below standard of care, or both. All of these factors contribute to the emergence of AMR. While human behavior contributes to AMR, another human endeavour—research innovation—provides a means to respond, for example, through the development of new antibiotics. The pace at which new antibiotics have been introduced has slowed considerably. For example, 16 antibiotics were approved by the FDA between 1983 and 1987, whereas only 2 were approved between 2008 and 2012, and a total of 5 new antimicrobials have been approved since the end of 2012. This slowdown is not unique to antibiotics; similar trends were observed for cardiovascular drugs and other agents. Nevertheless, certain characteristics of the antibiotic market likely hinder pharmaceutical industry investment in new drug development. Limited duration of treatment, relatively low prices per dose, the potential for the rapid emergence of resistance (and resulting uncertain market longevity), and antimicrobial stewardship efforts that limit access to new compounds all may curtail revenue prospects for a new antimicrobial agent.
_
Causes of AMR:
•Over-prescribing of antibiotics
•Patients not taking antibiotics as prescribed
•Unnecessary antibiotics used in agriculture
•Poor infection control in hospitals and clinics
•Poor hygiene and sanitation practices
•Lack of rapid laboratory tests
_
The level of antibiotic resistance is dependent on the following:
1. The population of organisms that spontaneously acquire resistance mechanisms as a result of selective pressure either from antibiotic use or otherwise,
2. The rate of introduction from the community of those resistant organisms into health care settings, and
3. The proportion that is spread from person to person.
All of these factors must be addressed in order to control the spread of antimicrobial-resistant organisms within health care settings. Community acquired antimicrobial resistance is increasing in large part because of the widespread suboptimal use of antibiotics in the outpatient settings and the use of antibiotics in animal husbandry and agriculture.
_
How antibiotic resistance evolves and spreads:
_
Causes of antimicrobial resistance can be divided into biological causes and societal causes:
Biological causes:
•Mutation: when microbes replicate themselves, genetic mutations can occur. Sometimes, these mutations can lead to the creation of a microbe with genes that aid it in surviving exposure to antimicrobial agents.
•Selective pressure: microbes that carry resistance genes survive to replicate themselves. The progeny of these resistant microbes will eventually become the dominant type.
•Gene transfer: microbes can also acquire genes from other microbes. Genes that have drug-resistant qualities are easily transferred between microbes easily.
Societal causes:
•Inadequate diagnostics: sometimes, a doctor will diagnose an infection without all the necessary information. This can lead to antimicrobials being prescribed “just in case,” or broad-spectrum antimicrobials being prescribed when a specific drug would be more apt. Both occasions accelerate AMR.
•Inappropriate use: if a course of antimicrobial drugs is not fully completed then some microbes may survive and develop resistance to the drug. If drugs are used for conditions they cannot treat then microbes can also develop resistance.
•Agricultural use: it is thought that more antibiotics are used in animal agriculture than for human diseases. The use of antibiotics in farm animals can promote drug resistance, and drug-resistant bacteria may be found on meat and in food crops exposed to fertilizer or water that is contaminated with animal feces, providing a route for animal to human transmission.
•Hospital use: critically ill patients are often given much higher doses of antimicrobials. The combination of more frequent utilization and close contact among sick patients creates an environment that is ideal for the spread of AMR germs.
______
Biocide abuse and antimicrobial resistance:
The term biocide includes disinfectants, antiseptics and preservatives. It does not include antibiotics, which, in spite of being biocides in the strictest sense, tend to be categorized separately. In recent years there has been a trend towards use of biocides in the home environment. These products have been marketed for decontamination of food preparation surfaces , areas perceived to be microbially contaminated (e.g. toilets) and general improvement of cleanliness in the home. Biocide resistance was first recognized nearly 70 years ago by Heathman et al. who identified chlorine resistance in Salmonella typhi, and antibiotic resistance was identified shortly after the availability of penicillin, but links between the two have only been recognized more recently. Because biocides tend to act concurrently on multiple sites within the microorganism, resistance is often mediated by non-specific means. Efflux pumps have the potential to act on a range of chemically dissimilar compounds and have been implicated in both biocide- and antibiotic-resistant bacteria. Cell wall changes may also play a role in the observed cross-resistance between biocides and antibiotics, probably by reducing permeability. Microbial changes that result in resistance to biocides and antibiotics should therefore cause concern. However, of equal significance, is the possibility of genetic linkage between genes for biocide resistance and those for antibiotic resistance.
_
McMurry & Levy reported in 1998 that mutations in the enoyl reductase gene (fab1) of Escherichia coli were associated with resistance to triclosan. This work suggested that Fab1 is the target for triclosan but failed to demonstrate any significant reduction in susceptibility to antibiotics in strains with fab1 mutations. However, the same workers have demonstrated that Inh1 (the mycobacterial analogue of the Fab1 protein) is a common target for triclosan and isoniazid in Mycobacterium smegmatis. Thus, it is possible that overuse of triclosan may select for antibiotic-resistant strains of mycobacteria. Mycobacterium tuberculosis, in contrast, is known to be intrinsically triclosan resistant but usually susceptible to isoniazid, and therefore this cross-resistance may not be of relevance to the majority of clinically important infections. Other workers have also demonstrated a link between biocide resistance and antibiotic resistance in atypical mycobacteria. Manzoor et al. demonstrated ethambutol resistance in strains of Mycobacterium chelonae that had been selected in vitro to be resistant to glutaraldehyde. This resistance was associated with changes in the composition of the cell wall, indicating that reduced permeability may be the mechanism for this cross-resistance. These links between biocide and antibiotic resistance are not confined to atypical mycobacteria, and it has been shown that resistance to the biocide benzalkonium chloride is closely linked to oxacillin resistance in Staphylococcus aureus. Akimitsu et al. reported that benzalkonium chloride-resistant mutants of methicillin-resistant S. aureus (MRSA) had oxacillin MICs as high as 512 mg/L, compared with 16 mg/L for the parent strain and 0.3 mg/L for methicillin-susceptible S. aureus (MSSA). Furthermore, resistance to both biocides and antibiotics can be plasmid mediated. A strain of gentamicin-resistant MRSA was investigated by Yamamoto et al. and was shown to contain a multidrug-resistant plasmid (pSAJ1) that conferred resistance to aminoglycosides, ethidium bromide, benzalkonium chloride and chlorhexidine. This plasmid, when transferred to E. coli, continued to express resistance to the same antibiotics and biocides as when in the original host. Plasmid-mediated resistance to biocides is a well-recognized phenomenon. Such resistance to quaternary ammonium compounds and other biocides has been identified in S. aureus, Pseudomonas spp. and members of the Enterobacteriaceae, and is mediated by specific genes (qacA, B, C, D and E). qacA, B and C (described in S. aureus) mediate resistance by an active efflux mechanism and have sequence homology with tetracycline efflux genes. qacE is a plasmid-mediated resistance gene found in Gram-negative organisms that also codes for an energy-dependent multidrug efflux mechanism. These resistance determinants are associated with resistance to a variety of antibiotics including trimethoprim, sulphonamides, oxacillin and aminoglycosides. Perhaps the most impressive example of biocide resistance that is linked to multiple antibiotic resistance is the mar regulon. Strains that constitutively express the Mar protein have over 60 chromosomal genes secondarily affected, and are resistant to tetracycline, chloramphenicol, triclosan and pine oil. It appears that such resistance to structurally unrelated compounds is mediated by an efflux mechanism.
_
The public is bombarded with advertisements advocating the use of biocides in the home. The implication is that homes are dangerous places, heavily contaminated with virulent microorganisms, and the only way to ensure the safety of one’s children is to use disinfectants liberally. This engenders a false sense of security in the public mind by suggesting that the widespread use of biocides reduces the organism load and thereby reduces the chance of acquiring infectious diseases. There are no data to support this stance; rather, as biocide use in the home environment as well as the healthcare environment continues to increase, the risk of selection of biocide-resistant strains must increase. Owing to the links between biocide resistance and antibiotic resistance outlined above there is a real risk that widespread biocide use could exacerbate the already worrying trend towards increased antimicrobial resistance in clinically relevant organisms. An essential part of preventing the spread of infection in the community and at home is proper hygiene. This includes hand-washing and cleaning shared items and surfaces. Antibacterial-containing products have not been proven to prevent the spread of infection better than products that do not contain antibacterial chemicals.
_
Antibacterial Soap v. Regular Soap:
The use of antibacterial soaps and detergents is no better at fighting germs than regular soap. It’s actually the friction of washing with soap and water that removes germ microbes. Rubbing the hands together breaks down the germ wall so that it washes down the drain. The type of soap used is less important. Some experts say that in a pinch, even just rubbing the hands on a paper towel for 20 seconds will remove some germs. The addition of an antibacterial agent, such as triclosan, to soap provides the added effect of killing organisms. But these products actually require prolonged contact to work — minutes, not seconds. This is why many antibacterials are useful in settings such as hospitals. At home, however, the average person doesn’t wash his hands long enough for triclosan to have the desired effect. It is recommended avoiding products with triclosan because of concerns over antibacterial resistance. These cautions stem from the fact residue from chemicals, such as, triclosan can stay on hands and surfaces at low concentrations and could lead to antibiotic resistance. FDA announced that it will not allow manufacturers of antibacterial hand soaps and body washes to market products that contain certain active ingredients. The agency has banned 19 ingredients in these products including triclosan and triclocarban—the two most commonly used ingredients.
_
In one study, researchers applied soap containing 150 micrograms of triclosan to a strain of wild (unmutated) E. coli — bacteria commonly found in raw food. The results showed that it took a full two hours for the agent to kill 90 percent of the bacteria. It took two to four times as much triclosan under the same conditions to kill 90 percent of a mutated strain. And adding antimicrobial chemicals to soap may be particularly ineffective. When researchers in the same study exposed the same strain of E. coli to just six micrograms of triclosan by itself, it killed as much of the bacteria in the same amount of time.
_
Study links Antibiotic Resistance with exposure to Chlorhexidine: a 2016 study:
Klebsiella pneumoniae bacteria exposed to chlorhexidine-containing disinfectants can become resistant to colistin, a last resort antibiotic often used against multidrug resistant pathogens. This is the first study to link exposure to chlorhexidine with resistance to colistin in this clinically important pathogen. The research is published in Antimicrobial Agents and Chemotherapy, a journal of the American Society for Microbiology. In the study, the investigators tested the hypothesis that K. pneumoniae could survive exposure to increased concentrations of chlohexidine, and that these exposures might cause resistance to commonly used antibiotics. They selected specific strains of K. pneumoniae that were representative of isolates routinely found in the clinic. Chlorhexidine is a common ingredient in a number of disinfectants used widely in the home and in healthcare settings, where it is a critical part of many infection control practices. While some strains died on exposure, others were able to survive at much higher concentrations of chlorhexidine than their parental strains. Some also gained resistance to colistin. The investigators also found gene mutations in the exposed K. pneumoniae that conferred resistance to both compounds. “Chlorhexidine is a critical part of current infection control practices, and the development of increased resistance to this compound has potential implications for our ability to prevent infections during routine and emergency surgery, and during admission to hospitals,” said coauthor J. Mark Sutton, PhD, scientific leader at the National Infections Service, Public Health England in Salisbury, UK. The study data, and other ongoing work in Sutton’s laboratory suggest that many bacterial pathogens may share the same, or similar metabolic pathways. Specific selective pressures from antiseptics, such as chlorhexidine, may result in similar mutations in these pathways conferring increased resistance in these different species of bacteria, he said. “If the same response is seen in hospitals, this might mean that we need to rethink how and where some types of critical disinfectants or antiseptics are used in the clinic.”
______
______
AMR in hospitals:
_
_
The figure below shows how AMR arises in hospital:
_
Risk factors for ARMs in hospital:
The following factors have been found to be associated with ARM colonization: underlying illness; intravenous, urinary, or enteral feeding devices; antibiotic use; wounds; decline in functional status; and increased intensity of nursing care. Irrational use of antibiotics, overdosing or under-dosing, self-medication and the inappropriate use of antimicrobials in a hospital setting are all to blame. One likely culprit is a lack of sanitation. In many areas, wastewater from hospitals is poorly filtered, allowing the antibiotic-resistant bacteria that flourish there to escape into waterways. If people drink this contaminated water or practise poor hygiene, the bacteria can spread. If you get increased antimicrobial use but don’t have the infrastructure for infection control, you’re setting yourself up for a tsunami of antibiotic resistance. Factors that favor the spread of ARMs in Long Term Care Facility include high resident to staff ratios, lack of attention to basic infection control measures, use of common equipment without disinfection between residents, limited facilities for handwashing and the inappropriate use of antimicrobials.
_
_
Hospital acquired infections (HAI):
Antimicrobial resistance is a predictable outcome of antimicrobial use. The rapidity with which resistance emerges and its extent are proportional to the intensity of antimicrobial use. Resistance first emerges in populations with a high frequency of infection, due to either underlying patient status or interventions compromising host defences, resulting in a high rate of antimicrobial use. Where patients at risk are in close proximity, the transmission of organisms between patients will be facilitated, and the opportunity for a single strain to disseminate widely is enhanced. All these features are present in health care facilities, particularly acute care facilities and areas such as intensive care units. Thus, health care facilities, particularly those which are large and care for the most complex patients, are a focal point in the emergence of antimicrobial resistance. The growing numbers of antimicrobial-resistant pathogens, which are increasingly associated with nosocomial infection, place a significant burden on healthcare systems and have important global economic costs. Effects include high mortality and morbidity rates, increased treatment costs, diagnostic uncertainties, and lack of trust in orthodox medicine. Despite a consensus that institutional infection control programs are important for containing antimicrobial resistance, the interactions between infection control activity and antimicrobial resistance are not straightforward. Optimal infection control programs, whose goal is to minimize nosocomial infections, may decrease the prevalence of resistance and infections caused by resistant organisms, but may also contribute to the emergence of antimicrobial resistance, and may be more effective in outbreak management because resistance facilitates identification of unusual organisms in the hospital. The overarching benefit of infection control programs in decreasing nosocomial infections, some of which may be with resistant organisms, is clear. The extent to which an intensification of infection control activity or expansion of responsibility to include containment of colonization with resistant organisms will benefit either the goal of decreasing nosocomial infections or decreasing endemic antimicrobial resistance cannot be estimated with information currently available. Promoting infection control activity to contain antimicrobial resistance in the absence of effective, highly restrictive, antimicrobial use programs would appear, ultimately, to be futile. Nonetheless, infection control programs in health care facilities should be supported and reinforced in their prime role—the prevention of infection, regardless of the presence or absence of antimicrobial resistance. This should lead to optimal patient outcomes, and limit the progression of resistance to the extent that infection control activity may have an impact.
Infection control in hospitals:
Standard precautions in hospitals are work practices that provide a basic level of infection control for the care of all patients, regardless of their diagnosis or presumed infection status. These precautions should be followed in all hospitals and healthcare facilities and include:
•good personal hygiene, such as hand washing before and after patient contact and the appropriate use of alcohol-based hand rub solutions
•the use of barrier equipment such as gloves, gowns, masks and goggles
•appropriate handling and disposal of sharps (for example, needles) and clinical waste (waste generated during patient care)
•aseptic techniques
Implementing standard precautions minimises the risk of transmission of infection from person to person, even in high-risk situations.
_
Rates of antimicrobial resistance in Gram-negative bacilli responsible for hospital-acquired infections:
| Study/surveillance network | INICC | SENTRY | ANSRPRG | EARS-NET |
| Geographic area | International (36 countries) | International (Europe/USA) | International (Asia) | International (Europe) |
| Study years | 2004–2009 | 2009–2011 | 2008–2009 | 2013 |
| Setting | ICU | ICU | ICU/non-ICU | ICU/non-ICU |
| Type of hospital-acquired infections | Catheter-related infections and ventilator-associated pneumonia | All (pooled) | Pneumonia | Bloodstream infections |
| Species/antimicrobial | ||||
| Escherichia coli | ||||
| Fluoroquinolones | 53% | 30% | – | 11–52% |
| 3GC | 67% | 13% | – | 5–40% |
| Carbapenems | 4% | <1% | – | 0–3% |
| Klebsiella pneumoniae | ||||
| Fluoroquinolones | – | 17% | 31% | 0–70% |
| 3GC | 72% | 19% | 43% | 0–70% |
| Carbapenems | 7% | 4% | 2% | 0–59% |
| Pseudomonas aeruginosa | ||||
| Fluoroquinolones | 45% | 30% | 30% | 0–53% |
| Aminoglycosides | 28% | 17%a | – | 0–51% |
| Piperacillin–tazobactam | 39% | 32% | 37% | 0–55% |
| Ceftazidime | – | 27% | 35% | 0–44% |
| Carbapenems | 45% | 30%b | 30% | 3–60% |
| Acinetobacter baumannii | ||||
| Ceftazidime | – | 63% | – | – |
| Carbapenems | 63% | 57%b | 67% | 0–90% |
ICU intensive care unit, 3GC third-generation cephalosporins.
aIndicator: gentamicin.
bIndicator: meropenem.
______
______
Antimicrobial overuse and misuse:
_
_
In the USA, an estimated 23 × 106 kg of antibiotics are currently used annually; about half are provided to people and the rest are manufactured for agriculture. In hospitals, they are generally administered parenterally, while in the community they are delivered mostly as oral preparations. About 7 × 106 kg of antibiotics, chiefly penicillins and tetracyclines, are used as growth promotants for food animals. Some 45 × 103 kg of antibiotics, namely tetracyclines and streptomycin, are provided as pesticides for agriculture; these are sprayed on to fruit trees in the southern and western USA. While this last amount seems small compared with overall antibiotic use, the geographical spread can be considerable. Some strains of Erwinia amylovora, the bacterial target of these drugs, have become resistant to antibiotics. While the emergence of resistant bacteria in agriculture is a small part of the overall global microbial resistance pool, it is an example of widespread antibiotic use in which the environment of microorganisms is besieged with growth-inhibitory agents. The result is the survival of those organisms that bear transposons and other mechanisms for self-preservation, leaving an environment of microorganisms that are largely resistant.
_
How are antibiotics provided? In some parts of the world, antibiotics are available over the counter in pharmacies, like other commodities. In some countries, vendors sell antibiotics on the street. This means of distribution provides the worst scenario for emergence of resistance: the possibility of too little drug for treatment and provision of drugs when not necessary. Of course, in industrialized areas like Europe and the USA, a prescription is required. But is that enough to assure prudent use? It appears not to be. A study was performed in Iran where a medical student, feigning a cold, entered 40 doctors’ offices and recorded physicians’ diagnosis and treatment. Thirty-seven of the forty physicians diagnosed a viral infection but still prescribed antibiotics. A few years ago, the New Yorker magazine aptly satirized American attitudes toward antibiotics with a cartoon of a doctor’s office sign stating, ‘Don’t forget to take a handful of our complimentary antibiotics on your way out’. According to the CDC, the prescription of antibiotics in outpatient settings could be reduced by over 30% without any adverse effects to patients’ health. A study by the CDC published in 2014 in the journal Pediatrics, found that 71% of cases of C. difficile identified in children aged 1-17 were community-associated, i.e. their infection was not associated with a hospital stay. What is more, 73% of these children had received a prescription for antibiotics in the 12 weeks prior to developing C. difficile, with most prescriptions made out in a doctor’s office. Also several studies have found that at least 50% of antibiotics prescribed in doctor’s offices for children are for respiratory infections, most of which do not require antibiotics. Official guidelines by the American Heart Association for dental antibiotic prophylaxis call for the administration of antibiotics to prevent infective endocarditis. Though the current (2007) guidelines dictate more restricted antibiotic use, many dentists and dental patients follow the 1997 guidelines instead, leading to overuse of antibiotics.
_
The figure below shows that countries using more antibiotics have higher AMR:
_
The overuse of antibiotics clearly drives the evolution of resistance. Epidemiological studies have demonstrated a direct relationship between antibiotic consumption and the emergence and dissemination of resistant bacteria strains. In bacteria, genes can be inherited from relatives or can be acquired from nonrelatives on mobile genetic elements such as plasmids. This horizontal gene transfer (HGT) can allow antibiotic resistance to be transferred among different species of bacteria. Resistance can also occur spontaneously through mutation. Antibiotics remove drug-sensitive competitors, leaving resistant bacteria behind to reproduce as a result of natural selection. Despite warnings regarding overuse, antibiotics are overprescribed worldwide. In the U.S., the sheer number of antibiotics prescribed indicates that a lot of work must be done to reduce the use of these medications. In many other countries, antibiotics are unregulated and available over the counter without a prescription. This lack of regulation results in antibiotics that are easily accessible, plentiful, and cheap, which promotes overuse. The ability to purchase such products online has also made them accessible in countries where antibiotics are regulated. The use of antibiotics in a population is the primary driver of the development of resistant bacteria. However, the factors underlying the development of resistance in pathogens is often more complex than simply using increasing amounts of a certain antibiotic. For certain pathogens, resistance to a particular antimicrobial is never seen. For example, group A streptococci have never developed resistance to penicillin; the reasons are unknown. Continued susceptibility of Treponema pallidum to penicillin indicates that resistance may not invariably occur despite powerful selective pressures that would favor its emergence. In spite of decades of extensive use of penicillin, group A streptococci remain exquisitely susceptible to this antibiotic barring exception of very few strains. This observation that continuing susceptibility has occurred despite the development of resistance to other antimicrobial agents demand explanation. Among the most likely explanations for this remarkable state of continued susceptibility to penicillin are that b-lactamase may not be expressed or may be toxic to the organism and/or that low-affinity penicillin-binding proteins either are not expressed or render organisms nonviable. Other potential explanations are that circumstances favorable for the development of resistance have not yet occurred and/or that there are inefficient mechanisms for or barriers to genetic transfer. It could be because despite millions of spontaneous random mutations, none could code for biochemical pathway to resist penicillin. It is because its genome is built in such a way that even millions of mutation cannot code for such pathway. I would call it genome incompatibility to resistance for a specific antibiotic. Also, the same factor of genome incompatibly to resistance prevents acquisition of penicillin resistance through plasmids. In other words, there are genetic factors in bacteria which prevent acquisition of specific resistance gene through plasmid. In other words, acquisition of resistance genes either by mutation or by plasmid is genetically correlated. This is a hypothesis which needs to be confirmed or rejected by experiments. If we could find genome code that confers incompatibility to resistance, we can introduce it in other bacteria so that they become sensitive to penicillin. Genome incompatibility to resistance for a specific antibiotic (exclusive sensitivity) is exactly opposite of intrinsic resistance due to innately coded genes which create natural “insensitivity” to a specific antibiotic. Of course ‘exclusive sensitivity’ is against evolutionary instinct to survive and therefore a ‘mistake’ evolutionary biologically. Time will tell whether a ‘mistake’ is corrected by converting ‘exclusive sensitivity’ to insensitivity.
_
AMR cycle:
Increased antibiotic use increase AMR which in turn increases antibiotic use.
_
Inappropriate Prescribing:
Incorrectly prescribed antibiotics also contribute to the promotion of resistant bacteria. Studies have shown that treatment indication, choice of agent, or duration of antibiotic therapy is incorrect in 30% to 50% of cases. One U.S. study reported that a pathogen was defined in only 7.6% of 17,435 patients hospitalized with community-acquired pneumonia (CAP). In comparison, investigators at the Karolinska Institute in Sweden were able to identify the probable pathogen in 89% of patients with CAP through use of molecular diagnostic techniques (polymerase chain reaction [PCR] and semiquantitative PCR). In addition, 30% to 60% of the antibiotics prescribed in intensive care units (ICUs) have been found to be unnecessary, inappropriate, or suboptimal. Incorrectly prescribed antibiotics have questionable therapeutic benefit and expose patients to potential complications of antibiotic therapy. Subinhibitory and subtherapeutic antibiotic concentrations can promote the development of antibiotic resistance by supporting genetic alterations, such as changes in gene expression, HGT, and mutagenesis. Changes in antibiotic-induced gene expression can increase virulence, while increased mutagenesis and HGT promote antibiotic resistance and spread. Low levels of antibiotics have been shown to contribute to strain diversification in organisms such as Pseudomonas aeruginosa. Subinhibitory concentrations of piperacillin and/or tazobactam have also been shown to induce broad proteomic alterations in Bacteroides fragilis. The problem of resistance is exacerbated by a wide range of fixed-dose combinations in the market, often without scientific or medical merit or evaluation. A recent study reported 48 fixed dose combinations and 22 loose antimicrobials for tuberculosis. Loose antimicrobials come without packaging and do not mention the name of the drug, its manufacturer, the date of manufacture, or the date of expiry. There is poor clinician awareness of the rationality and dosing of fixed-dose combinations.
__
Antibiotics Fight Bacteria, Not Viruses:
Antibiotics are meant to be used against bacterial infections. For example, they are used to treat strep throat, which is caused by streptococcal bacteria, and skin infections caused by staphylococcal bacteria. Although antibiotics kill bacteria, they are not effective against viruses. Therefore, they will not be effective against viral infections such as colds, most coughs, many types of sore throat, and influenza (flu). Antibiotics have no effect on viral infections such as the common cold. They are also ineffective against sore throats, which are usually viral and self-resolving. Most cases of bronchitis (90–95%) are viral as well, passing after a few weeks—the use of antibiotics against bronchitis is superfluous and can put the patient at risk of suffering adverse reactions.
Using antibiotics against viral infections:
•will not cure the infection
•will not keep other individuals from catching the virus
•will not help a person feel better
•may cause unnecessary, harmful side effects
•may contribute to the development of antibiotic-resistant bacteria
A 2015WHO survey across 12 countries found that 64% of the public wrongly believe that antibiotics also work for viral infections such as influenza and colds. Such basic knowledge gaps lead patients and physicians to reach for antibiotics without appreciating the costs of antimicrobial resistance. Patients and health care professionals alike can play an important role in combating antibiotic resistance. Patients should not demand antibiotics when a health care professional says the drugs are not needed. Health care professionals should prescribe antibiotics only for infections they believe to be caused by bacteria. As a patient, your best approach is to ask your health care professional whether an antibiotic is likely to be effective for your condition. Also, ask what else you can do to relieve your symptoms.
_
Physicians:
Physicians influence antimicrobial use in three distinct ways: by giving recommendations to obtain antimicrobials (unwritten or ‘verbal’ prescriptions), by issuing written prescriptions or by prescribing and directly dispensing/selling drugs. Physicians’ prescribing habits may be an important determining factor in ‘self-medication’, as they are often seen as authority figures and their written prescriptions are valued. According to Hardon (1991), doctors may even have a role of ‘legitimising’ popular choices of pharmaceuticals, and the consequences of their antimicrobial prescribing practices may extend to other actors. In India, 75% of pharmacy clients based their antimicrobials purchasing decision on an earlier prescription by a physician (Dua et al. 1994). In Mexico, antimicrobial therapy was found to be up to seven times more likely if a sick person had seen a physician (Bojalil and Calva 1994). Furthermore, in a Mexican study, 61% of all episodes of diarrhoea were treated with one or more antimicrobials that had been prescribed by a physician (Bojalil and Calva 1994). Similar findings are reported from other countries (Lansang et al. 1990; Calva 1996; Bartoloni et al. 1998).
_
Physicians’ knowledge of correct prescribing is a fascinating subject. A number of papers have shown that prescribers may demonstrate correct knowledge, but that they may still practice differently. This suggests that other determinants are stronger. Some of these determinants have been explored in the literature, but much remains to be explored. The often heard assumption that lack of quality diagnostic services drives poor antimicrobial use remains to be proven, as even when laboratory facilities are available, prescribers may not use them. Prescribers often reported a fear of poor disease outcomes without using antimicrobial agents. This fear may link to the powerful image of antimicrobials and the notion that without prescribing them, not all clinical options have been tried out. Reports give examples of how doctors deal with insecurity on diagnosis and treatment, but it continues to be unknown whether these fears can be addressed in other ways than applying antimicrobial agents. On the other hand, as antimicrobials are often perceived as risk-free agents, doctors may feel that there are few arguments not to prescribe an antimicrobial agent. Risk perceptions of prescribers and how these can be used to improve practices of antimicrobial use are largely unexplored. Peer norms and local medical cultures impress as important determinants of prescribing by doctors, pharmacist dispensing and community use. Interestingly, few studies have investigated the role of peer influence in inappropriate use of antimicrobials. Determinants of antimicrobial use are complex, involving a variety of motivations on the part of prescribers, dispensers and consumers. Systematically expanding our knowledge about factors that determine antimicrobial use, their responsiveness to change, and cost-effective strategies for achieving these changes, will be indispensable to effectively counteract the rise of resistance.
_
Availability of Laboratory Services:
Lack of access to quality laboratory services is often regarded as a deterrent to rational use of antimicrobials. In Bangladesh, more than 90% of antimicrobials were reported to be used without establishing a laboratory diagnosis. In Trinidad, physicians explained that they do not normally request laboratory investigations, as they consider them unnecessary and the waiting time for results is felt to be too long (Mohan et al. 2004). Similarly, lack of laboratory facilities, or the inability of patients to pay for such services, was blamed for over-prescribing of antimicrobials in a Pakistani study (Nizami et al. 1996). An attractive alternative to using laboratory services may be to use antimicrobials as a diagnostic tool. When a patient does not recover after an initial antimicrobial treatment, further diagnostic activities may be employed (Hardon 1991). Interestingly, the availability and accessibility of laboratory facilities and personnel does not necessarily motivate physicians to use them. All hospitals in a Malaysian study had facilities for microbiological culture, but only 20% of antimicrobial prescriptions were made on the basis of microbiological reports (Lim and Cheong 1993). Javato-Laxer and colleagues (Javato- Laxer et al. 1989) explained that because of their failure to determine aetiologies of infections, physicians often prefer to use broad-spectrum antibiotics, believing that this will cover all possible aetiologies and unusual pathogens.
_
Pressure of Pharmaceutical Promotion:
Company sales representatives and commercially oriented drug publications are known to be an important source of information for prescribers (Bosu and Ofori-Adjei 1997). The few studies that carefully followed and interviewed pharmaceutical representatives (Kamat and Nichter 1987; Wolffers 1987) concluded that promotional activities increased the haphazard supply of antimicrobials in some societies. In Indonesia, prescribers receive payment for issuing certain drugs during promotional events. In the Philippines, pharmaceutical companies reinforce the notion of risk-free medicines and promote a ‘why worry’ attitude among doctors (Van Staa 1993). Nevertheless, reports on commercial pressures in drug prescribing in developing countries tend to be anecdotal, and understanding of this influence is limited.
__
Several studies link antibiotic use to antibiotic resistance:
_
Study-1
Antibiotics are also fed to animals. In the mid-1970s, authors performed a study that involved raising 300 chickens on a small farm outside Boston. They provided 150 newly hatched chicks with oxytetracycline-laced feed and another 150 without. They followed the effect of the antibiotic-laced feed on the animals and people on the farm. As they began the study, the control group had little or no resistant organisms. In the group receiving low levels (200 ppm) of oxytetracycline, tetracycline resistance began to emerge among the faecal Escherichia coli. What was surprising was that, within 12 weeks, they detected as much as 70% of all E. coli with resistance to more than two antibiotics, including ampicillin, sulphonamides and streptomycin. The resistances were all on transferable plasmids that emerged following use of just tetracycline.
_
Study-2
Treating Acne with Antibiotics leads to Resistance: a 2001 study:
The use of antibiotics to treat severe acne can lead to the development of antibiotic resistance in the bacteria that cause the skin condition, say researchers from the Karolinska Institute in Stockholm, Sweden. They reported the results of their study at the 101st General Meeting of the American Society for Microbiology (ASM) in Orlando, Florida. “Antibiotic-resistant Propionibacterium acnes (the bacterium that causes acne) strains were significantly more often isolated from antibiotic-treated than from non-antibiotic-treated patients. When patients with acne are treated with antibiotics, the risk of development of antibiotic resistance should be realized,” says Carl Erik Nord, the lead investigator of the study. P. acnes is part of the natural microflora of the skin and is thought to play an important role in the development of severe, inflamed acne. Antibiotics, most commonly tetracycline and erythromycin, have been used for over 20 years to treat moderate and severe forms of acne. The use of antibiotics has been connected to the development of antibiotic resistance in other bacteria, and the researchers sought to determine if that was also occurring in P. acnes.
_
Study-3
High doses of antibiotics may have potential to promote increased cross-resistance: a 2014 study:
An experimental evolution approach has been used by researchers to evolve 88 different E. coli populations against 22 antibiotics, under ‘strong’ and ‘mild’ selection conditions. Results demonstrate that the evolution of cross-resistance depends on selection strength. Overall, they found evidence for higher cross-resistance in the strongly selected strains and higher numbers of pathway-specific mutations. The study yielded important new insights into the increased emergence of drug resistance with the use of high doses of antibiotics, as well as hypersensitivities to exploit for new antibiotic therapies.
_
Study-4
Prolonged Antibiotic Prophylaxis after Cardiovascular Surgery and Its Effect on Surgical Site Infections and Antimicrobial Resistance: a 2000 study:
Antimicrobial prophylaxis complements meticulous technique in reducing the incidence of surgical site infections after cardiovascular surgery. Although the principles of surgical prophylaxis have been outlined on numerous occasions, there is still misuse of antimicrobials for this purpose. The results demonstrate that prolonging antibiotic prophylaxis beyond 48 hours after CABG surgery is still frequently practiced but does not decrease the risk of surgical site infections. Moreover, it results in an increased risk of acquired antibiotic resistance and should therefore be avoided.
______
The Pharmacological Side of Resistance:
Two pharmacological issues have particular relevance to the resistance problem:
(a) spectrum and (b) compliance.
a) Wide-spectrum antibiotics do have an important role in fighting infection, either those caused by several different bacterial species or those for which assessing the etiology is too difficult or takes too much time. However, wide spectrum has been an obvious goal in the R&D of antibiotics, as it ensures also a wide variety of clinical uses and, of course, of sales. Wide spectrum has also been presented to the medical community as a general advantage so that the physician need not worry about the etiology of an infectious disease to start treatment. Of course, the likelihood of this strategy to succeed in the short term and the individual patient is high. But this shotgun notion contributes to resistance as it applies selective pressure, not only upon the etiological agent of the infectious episode but also upon a larger fraction of the patient’s microbiota. Some reports indicate that, although worldwide use of antibiotics is receding, the use of wide-spectrum ones is dramatically increasing. Along with increasing resistance trends, marketing efforts are aimed at positioning newer fluoroquinolones as choice drugs against lower respiratory tract infections, instead of aminopenicillins and macrolides, and even oral, third generation cephalosporins against diseases as minor as pharyngotonsillitis. It is of course not surprising to see among community-acquired uropathogens up to one-third of them highly resistant to fluoroquinolones, and even a high prevalence of ESBL enzymes, previously confined to hospital settings.
b) Compliance is a pharmacological issue that has many repercussions upon bacterial resistance. Patients often miss drug doses, both by mistake and deliberately (there is, for instance, the urban legend that antibiotics and alcohol interact dangerously so that people under antibiotic therapy, but attending a party, would rather miss a dose than avoid alcohol at the party). Other patients decide to suspend the treatment prematurely when they feel well enough. Both instances result in the exposure of surviving pathogens to subinhibitory concentrations of antibiotics and, consequently, to increased chances of acquiring resistance. When a patient gets antibiotics through self-prescription, the patient is more likely to take shorter treatments. A side effect of a lack of compliance is to have remaining doses of antibiotics that are often stored ‘‘just in case’’; this favors future self-prescription, using antibiotics even beyond their expiration dates, which could expose bacteria to further subinhibitory concentrations. A list of suggestions for improving patient compliance is included in Goodman & Gilman’s Pharmacological Basis of Therapeutics, which is sound for the United States and other developed countries, but is simply unrealistic for poor countries where overworked physicians cannot devote the time needed for approaches such as ‘‘developing satisfactory, collaborative relationships between doctor and patient. . .’’ and ‘‘using behavioral techniques such as goal setting, self-monitoring, cognitive restructuring. . .’’ and where ‘‘easy-to-read written information’’ is not useful for an illiterate patient, while ‘mechanical compliance aids’ are simply not available. However, it might be useful to try to dispel wrong notions (such as the alcohol–antibiotic interaction; but, then again, the notion might have been acquired from misinformed physicians) and to teach the patient that the risk of not taking each and every dose is not only – or perhaps not even – for him/her but for his/her family, friends, and coworkers, in the mid- and long term (but, then again, physicians would need to be convinced of these themselves).
_
Consequences of Empirical Antibiotic Treatment:
Empirical treatments are experienced-based, therapeutic regimens generally administered prior to confirmatory diagnosis. Resistance may complicate early treatment of infection before culture results are known, such as with empirical therapy for community-acquired pneumonia or urinary tract infections, as well as for surgical prophylaxis. For example, fluoroquinolones have long been used as perioperative prophylaxis for patients undergoing transrectal prostate biopsies. In the past decade, increasing reports of postprocedure sepsis with fluoroquinolone-resistant E coli have led to new preprocedure selective rectal cultures for such strains, which require an additional visit to the urologist and specialized microbiology testing. Alternatively, some hospitals have broadened surgical prophylaxis to drug combinations such as ceftriaxone and gentamicin that cover fluoroquinolone-resistant E coli. However, this approach may select for different resistant strains because of the broad-spectrum coverage, thereby delaying the clinical consequences of resistance until a later date.
__
Role of patient/ consumer in AMR:
Self-medication:
Several authors have argued that ‘self-medication’ is the key determinant of improper antimicrobial use (Obaseiki-Ebor et al. 1987). As consumers are believed not to have correct knowledge on antimicrobial use, ‘self-medication’ is assumed to be undesirable. ‘Self-medication’ is practised widely, and appears to be by far the most common medical response (Van der Geest 1982; Hardon 1991). In a community study in Brazil, antimicrobials were the group of medicines that were most often used in ‘self-medication’ (Haak 1988). In a Nigerian community, all members admitted having used an antimicrobial in recent times, and most of them admitted that they had treated themselves once or more with antimicrobials before consulting a physician (Obaseiki-Ebor et al. 1987). Surveys of pharmacy sales confirm high rates of ‘self-medication’ with antimicrobials. Motives for ‘self-medication’ with antimicrobials include the need to save money, and the desire to treat ‘confirmed’ or suspected bacterial infections.
_
Patients’ Interviews and Misuse of Antibiotics: a 2001 survey:
To better evaluate patient contribution in antibiotic use, authors questioned 5379 subjects from 9 countries. Antibiotics are perceived as strong, efficient drugs, but they are believed to undermine immunity. Interviewees believe that most respiratory infections, except the common cold, require antibiotic therapy and 11% of them had to exaggerate their symptoms to get an antibiotic prescription from their physician. About 1 patient in 4 saved part of the antibiotic course for future use. Sixty-nine percent of the patients claimed to have taken the course until the end (United Kingdom, 90%; Thailand, 53%), and 75% claimed that they actually took all the daily doses. In all countries, it was possible to get antibiotics from a pharmacist without a medical prescription. This study shows that patients exert pressure on their doctors to get antibiotics and should allow a design for precise educational action aimed at the public for better control of antibiotic use in the community. This survey suggests that patients represent a significant source of antibiotic misuse in the community. Authors conclude this study by highlighting the need to educate patients regarding antibiotic use and the consequences of misuse: what diseases actually require antibiotics, why full daily doses must be respected, absence of significant alterations of immunity associated with antibiotic therapy, danger of keeping part of a course for future uncontrolled use, and need of a prescription for getting antibiotics from the pharmacist could be some of the issues to be discussed with the patients.
_______
Pharmacy, quackery and pharmaceutical companies:
_
Doctors, unlicensed medical practitioners and illicit drug sales are the main culprits behind antibiotic resistance in India, the country’s agriculture ministry said in a statement that shifted blame for the growing public health problem away from veterinarians. The use of antibiotics in veterinary medicine “is not a sole cause of drug resistance,” said S.K. Dutta, assistant commissioner of the department of animal husbandry in the Ministry of Agriculture and Farmers Welfare. Traditional medical practitioners generally practice without formal supervision. They often lack access to medical technology or other diagnostic services and they rarely receive training in antimicrobial prescribing. Their information on indications, contraindications and side effects of drugs tends to come from informal, non-medical sources or sometimes from pharmaceutical representatives. Some traditional healers apply western medicine, including the use of antimicrobials (Wolffers 1987; Singh and Raje 1996). Competition with western doctors may be stiff and the ability to prescribe antimicrobials is believed to attract patients. For example, an ayurvedic healer in India used penicillin injections in the treatment of serious infections, such as skin ulcerations, pulmonary tuberculosis, abscesses and conjunctivitis. Because his patients demanded antimicrobials, the ayurvedic healer was unable to eliminate penicillin from his practice (Burghart 1988).
_
Antibiotics being handed out ‘like smarties’ by online pharmacies:
A BBC investigation has revealed a rise in the number of online pharmacies selling antibiotics without face-to-face consultations. A number of medical organisations, including the British Dental Association (BDA), have raised concerns over the way antibiotics can be prescribed to patients without any checks and against National Institute for Health and Care Excellence (NICE) guidelines. This BBC investigation highlights an urgent need to raise public awareness of AMR, and the importance of seeing a dentist if people are concerned they might have a dental infection, rather than going online for antibiotics. Faye Kirkland, who undertook the BBC investigation, was able to get three different antibiotics from an online store for conditions, including a dental problem, within 24 hours. The health risks presented by AMR require a change in gear from patients, practitioners, and policymakers.
__
Pharmacies:
Pharmacies are often the first source of advice for patients who seek care (Lansang et al. 1990; Hui et al. 1997). In most developing countries pharmacists are legally barred from diagnosis and prescription, but in practice they often do. They may even have special rooms for physical examinations and injections (Serkkola 1990; Van Staa 1993). In some instances they have commercial contacts with medical clinics or private doctors. Pharmacy personnel in the Philippines and Mexico give patients advice to buy antimicrobials (Lansang et al. 1990; Calva 1996), while in Egypt pharmacy staff simply refill old bottles of antimicrobials, in most cases, without asking for a prescription (Khallaf et al. 1991). Mothers in poor Brazilian urban slums were reported to seek treatment from pharmacies because it is cheaper and less time-consuming (Schorling et al. 1991).
_
Pharmaceutical Industry Influence:
Influence of the pharmaceutical industry is often mentioned in papers dealing with medicine use. Reports from Brazil mentioned pharmacies trying to sell more of certain drugs because of incentives provided by suppliers (Haak 1988), and a report from the Philippines (Van Staa 1993) describes how pharmaceutical representatives visit pharmacies and physicians to boost consumption of defined drugs. In Sri Lanka, pharmacy attendants admitted that sales representatives were their major source of information on drugs (Wolffers 1987). Apparently the pharmaceutical industry is able to change drug consumption patterns, but how this influence works seems to be less well investigated and known. Attention has been called for this lack of information on industry marketing practices in developing countries (Van der Geest et al. 1996), and applying ‘marketing’ methods may offer options that have been overlooked so far.
_
Availability of Few New Antibiotics:
The development of new antibiotics by the pharmaceutical industry, a strategy that had been effective at combating resistant bacteria in the past, had essentially stalled due to economic and regulatory obstacles. Of the 18 largest pharmaceutical companies, 15 abandoned the antibiotic field. Mergers between pharmaceutical companies have also substantially reduced the number and diversity of research teams. Antibiotic research conducted in academia has been scaled back as a result of funding cuts due to the economic crisis.
The figure above shows number of antibacterial New Drug Application approvals versus Year Intervals:
_
Antibiotic development is no longer considered to be an economically wise investment for the pharmaceutical industry. Because antibiotics are used for relatively short periods and are often curative, antibiotics are not as profitable as drugs that treat chronic conditions, such as diabetes, psychiatric disorders, asthma, or gastroesophageal reflux. A cost–benefit analysis by the Office of Health Economics in London calculated that the net present value (NPV) of a new antibiotic is only about $50 million, compared to approximately $1 billion for a drug used to treat a neuromuscular disease. Because medicines for chronic conditions are more profitable, pharmaceutical companies prefer to invest in them. Another factor that causes antibiotic development to lack economic appeal is the relatively low cost of antibiotics. Newer antibiotics are generally priced at a maximum of $1,000 to $3,000 per course compared with cancer chemotherapy that costs tens of thousands of dollars. The availability, ease of use, and generally low cost of antibiotics has also led to a perception of low value among payers and the public. In addition, microbiologists and infectious-disease specialists have advised restraint regarding antibiotic use. Therefore, once a new antibiotic is marketed, physicians—rather than prescribing it immediately—often hold this new agent in reserve for only the worst cases due to fear of promoting drug resistance, and they continue to prescribe older agents that have shown comparable efficacy. Therefore, new antibiotics are often treated as “last-line” drugs to combat serious illnesses. This practice leads to the reduced use of new antibiotics and a diminished return on investment. When new agents are eventually used, the emergence of resistance is nearly inevitable. However, since bacterial evolution is uncertain, the timeline for the development of resistance is unpredictable. A manufacturer that invests large sums of money into antibiotic development may therefore discover that profits are prematurely curtailed when resistance develops to a new antibiotic. Economic uncertainty related to the Great Recession has also had a restraining effect on the end users of antibiotics. Developed countries with well-funded health care systems have applied austerity measures, while developing countries such as China and India still have a large cohort of population that cannot afford expensive new medicines. As an additional complication, most antibiotics are currently off-patent and are supplied by manufacturers of generic drugs. The result has been access to cheap and generally effective drugs, which is good for the public; however, the downside is that many payers expect all antibiotics to be priced similarly—even new agents that target multidrug-resistant (MDR) pathogens. Because of these factors, many large pharmaceutical companies fear a potential lack of return on the millions of U.S. dollars that would be required to develop a new antibiotic. The Infectious Diseases Society of America (IDSA) reported that as of 2013, few antibacterial compounds were in phase 2 or 3 development. In particular, the IDSA noted that unacceptably few agents with activity against emerging, extensively resistant gram-negative bacteria, such as Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter baumannii, were being developed. Pharmaceutical companies have also taken a more active interest in developing antibiotics for methicillin-resistant Staphylococcus aureus (MRSA), rather than gram-negative pathogens. The most likely explanation for this imbalance is that MRSA is a major problem worldwide, whereas the market for treating gram-negative organisms is smaller, and somewhat more unpredictable given that resistance is rapidly acquired.
_
Backup treatments can have serious side-effects; for example, treatment of multi-drug-resistant tuberculosis can cause deafness or psychological disability. The potential crisis at hand is the result of a marked decrease in industry R&D. Poor financial investment in antibiotic research has exacerbated the situation. The pharmaceutical industry has little incentive to invest in antibiotics because of the high risk and because the potential financial returns are less likely to cover the cost of development than for other pharmaceuticals. In 2011, Pfizer, one of the last major pharmaceutical companies developing new antibiotics, shut down its primary research effort, citing poor shareholder returns relative to drugs for chronic illnesses. However, small and medium-sized pharmaceutical companies are still active in antibiotic drug research. Developing new antibiotics is an obvious solution to the problem of drug-resistant bacteria but it isn’t happening. Why not? The drug companies don’t see much profit in it. Developing a new antibiotic and getting it approved for therapeutic use takes a lot of time and money. If you are successful, you may save a lot of lives but you won’t make a lot of money because if the antibiotic works, the patient stops needing it and stops buying it. Drug companies would much rather make drugs for chronic conditions that people are told to buy for months or years or lifetimes.
_
Why no new antibiotics?
It sounds simple, doesn’t it? If bacteria are becoming resistant to current antibiotics, why not just develop new ones? But developing a new antibiotic doesn’t happen overnight. In fact, there have been no wholly new antibiotics in over a decade. There are several reasons for this.
1. Economic:
It may sound callous, but from a commercial point of view, there’s really not that much money to be made from antibiotics. In 2015, only six of the world’s top 50 pharmaceutical companies are still pursuing antibiotic research. After all, why would major pharmaceutical companies invest time and money developing drugs that are only taken for a short time to achieve a cure, when they could be developing drugs to treat chronic illness (such as high blood pressure or cholesterol) that are taken for life? In addition, any new antimicrobial will inevitably have its usefulness reduced by the development of resistance in target micro-organisms.
As you can see that only after 20 years, the pharmaceutical company can break even when new antibiotic is researched.
_
2. Scientific:
Anitmicrobials are complex and need very large investments in research and development before they can be marketed. Most never make it to market and many companies state the scientific challenges of antibiotic development as disincentives to continue further research. One oft-repeated line is that the ‘low hanging fruit has been picked’, which basically translates as ‘the easy or basic antibiotics have been found, and now it’s much harder to find and develop new ones’. Indeed, drug screens for new antibiotics tend to re-discover the same lead compounds time and again. Thus, discovery and development of antibiotics has become scientifically more complex and more time consuming—and therefore more expensive. Taking the baton from the large pharmaceutical companies, small and medium biotech firms have become the innovators in the industry, working to find new antimicrobials which they hope will then be acquired by their larger counterparts. At the end of the day however, money and profit is still the key driver in this industry. In the decades after penicillin came to market drug companies fell over each other to develop new antibiotic molecules. Since then, interest has waned. The pipeline of potential new products at various stages of clinical trial is barely 40 strong. Only a fraction of them will reach market as seen in the figure below; each will represent a big investment.
_
3. Regulatory:
Getting a new drug approved is never easy. There are multiple regulatory requirements that companies must manoeuvre and red tape that must be cleared. The difficulty of the process, coupled with the scientific and economic considerations, have resulted in many antimicrobials being placed in the ‘too hard’ basket. To combat the lack of interest from major companies, the US Government has stepped in with public incentives, patent protection, fast-track approval processes and partnerships to try to promote interest in these ‘forgotten drugs’. In 2015 President Barack Obama dedicated US$1.2 billion from the annual budget to fight against AMR infections. It is hoped that this investment may go some way towards reviving interest in antibiotic development.
_
The consequences of the failure to create new antibiotics could be catastrophic. Availability of effective antibiotics has revolutionized public health and has been responsible for enabling countless advancements in medical care. For example, antibiotics have been critical to the development of advances in surgery and of myeloablative therapies for cancer and to the transplantation of both solid organs and hematopoeitic stem cells. Effective antibiotics have also been critical for advanced medical treatment of patients with trauma and battlefield injuries, as well as myocardial infarctions, strokes, and other illnesses that require intensive care with catheters, hyperalimentation, and mechanical ventilation. Ironically, the very advances in medical care enabled by effective antibiotic therapies have, in turn, created enormous populations of increasingly immunocompromised hosts, who develop infections caused by increasingly resistant microbes that require treatment with newer, more powerful antibiotics. As global and US populations continue to age, this upwardly spiralling need for intensive care with catheters and ventilators, for increasingly aggressive cancer chemotherapy, and for cardiac, abdominal, and other complicated surgeries are all going to continue to increase. Although we have come to take for granted such elements of modern medical care, their continued utility depends in large part on the continued availability of effective antimicrobial therapy.
__
At the end of the day, after so many years of research and clinical trials, when a new antibiotic comes to market, it may become ineffective due to AMR. The figure below shows how a new wonder antibiotic gets undermined by antimicrobial resistance:
______
_______
Antimicrobial use in animals:
_
_
As global demand for animal protein grows, antibiotics are increasingly used to raise food-producing animals in intensive production – mostly to promote growth, rather than treat disease. The result is an increasing prevalence of antibiotic-resistant bacteria in livestock, poultry and aquaculture. Also many farmers are transitioning to intensive agriculture and often use antibiotics to optimise production. More antibiotics are used in poultry, swine and cattle to promote growth and prevent disease than are used by the entire human population. Globally, livestock consumed at least 63,200 tons of antibiotics in 2010, accounting for nearly 66% of the estimated 100,000 tons of antibiotics produced annually worldwide, which is projected to rise to 105,600 tons by 2030. In 2010, China was estimated to consume the most antibiotics in livestock, followed by the United States, Brazil, Germany, India, Spain, Russia, Mexico, France and Canada. The same antibiotics then find their way into municipal water systems when the runoff from housing facilities and feedlots contaminates streams and groundwater. So it’s a double hit: we get antibiotics in our food and drinking water, and we meanwhile promote bacterial resistance. Routine feeding of antibiotics to animals is banned in the European Union and many other industrialized countries. The World Health Organization concluded that inappropriate use of antibiotics in animal husbandry is an underlying contributor to the emergence and spread of antibiotic-resistant germs, and that the use of antibiotics as growth promoters in animal feeds should be restricted.
_
Antibiotic use in livestock:
_
Eighty percent of antibiotics sold in the United States are used on livestock. The majority of these antibiotics are given to animals that are otherwise healthy. Rather, it is normal practice to mix antibiotics with livestock food to promote healthier living conditions and to encourage animal growth. The use of antibiotics in animals is to a large degree involved in the emergence of antibiotic-resistant microorganisms. Antibiotics are used in food with the intention of not only preventing, controlling, and treating diseases, but also to promote growth. Antibiotic use in animals can be classified into therapeutic, prophylactic, metaphylactic, and growth promotion uses of antibiotics. All four patterns select for bacterial resistance, since antibiotic resistance is a natural evolutionary process, but the non-therapeutic uses expose larger number of animals, and therefore of bacteria, for more extended periods, and at lower doses. They therefore greatly increase the cross-section for the evolution of resistance. Demand for animal protein for human consumption is rising globally at an unprecedented rate. Modern animal production practices are associated with regular use of antimicrobials, potentially increasing selection pressure on bacteria to become resistant.
_
Growing antibiotic use in the animal sector is resulting in a greater selection of pathogens and is being driven by increased demand for meat and poultry. The extreme growth in consumption of chickens is primarily the result of the expansion of this sector in India alone, where areas of high consumption (30 kg per km2) are expected to grow 312% by 2030. A recent Organisation for Economic Co-operation and Development (OECD) report indicated that the costs of withdrawing antimicrobial growth promoters in India would be roughly US$1.1 billion. However, widespread resistance may hold more consequence for India than for other countries because of India’s high bacterial disease burden. Currently, India does not have regulatory provisions for the use of antimicrobials in cattle, chickens, and pigs raised for domestic consumption. Recent studies in various regions of India have discovered antimicrobial residues in food animal products (such as chicken meat and milk), indicating that antibiotic use in food animal production is widespread.
_
The figure above shows origins of antibiotic-resistant Staphylococcus aureus :
_
_
AMR transmission from animals to humans:
In research studies, occasional animal-to-human spread of antibiotic-resistant organisms has been demonstrated. Resistant bacteria can be transmitted from animals to humans in three ways: by consuming animal products (milk, meat, eggs, etc.), from close or direct contact with animals or other humans, or through the environment. In the first pathway, food preservation methods can help eliminate, decrease, or prevent the growth of bacteria in some food classes. Although conclusive evidence directly linking the use of drugs in food animals to an increase in drug-resistant bacteria that make people sick has not been uncovered, a number of recent studies suggesting such a link concern many scientists. “There is no evidence that antibiotic resistance is not a problem, but there is insufficient evidence as to how big a problem it is,” says Dr. Margaret Mellon, with the Union of Concerned Scientists. In one study published in the New England Journal of Medicine on February 6, 2002, researchers found links that strongly suggested that the people who developed Cipro-resistant bacteria had acquired them by eating pork that were contaminated with salmonella. The report concluded that salmonella resistant to the antibiotic flouroquine can be spread from swine to humans, and, therefore, the use of flouroquinolones in food animals should be prohibited. Another New England Journal of Medicine study from Oct. 18, 2001, found that 20 percent of ground meat obtained in supermarkets contained salmonella. Of that 20 percent that was contaminated with salmonella, 84 percent was resistant to at least one form of antibiotic. Antimicrobial resistance due to a particular antibiotic used in food animals may result in reduced efficacy of most or all members of that same antibiotic class, some of which may be extremely important for human medicine. This occurs because of the similarity of the antibiotic’s related structural components, which causes cross-recognition and cross-resistance for all or most of the antibiotics within the same antibiotic class. An example is the emergence and spread of vancomycin resistant enterococci (VRE) in hospitals following the extensive use of avoparcin in animals, a glycopeptide antimicrobial agent that is structurally similar to vancomycin. Another example is virginiamycin resistance cross-reacting with resistance to the human streptogramin, quinupristin-dalfopristin. Acquisition of resistant bacteria from farm animals has been shown to occur either via ingestion of foods of animal origin or via direct contact with infected animals. Because of their survival advantage, resistant bacteria may remain viable for longer periods in the environment and in animal reservoirs where they can eventually be transmitted to humans. There is evidence that transmission of MRSA strains can occur from animals to humans, and vice-versa. MRSA has been found in humans closely associated with carrier animals; among pet owners, veterinarians and veterinary personnel as well as pig and cattle farmers. Studies identified both livestock and companion animals as potential sources of MRSA for humans, and close contact with these animals was identified as a risk factor for their carriage in people. Recent Indian study suggests that chickens could be a major source for transmission of emerging MDR pathogen, H. pullorum, from poultry to humans. Additionally the study strongly supports the hypothesis that this species is an emerging pathogen, as it is closely related to established pathogens such as Campylobacter jejuni. Given the prevalence of H. pullorum in Indian chicken, as described in the study, the half billion Indians who eat chicken, and the fact that chicken consumption is growing at a huge 12 percent per year, the potential for spreading multi-drug resistance is alarming. Animals can serve as mediators, reservoirs and disseminators of resistant bacterial strains and/or AMR genes. Consequently, inappropriate use of antimicrobials in animals may eventually result in increased human morbidity, increased human mortality, reduced efficacy of related antibiotics used for human medicine, increased healthcare costs, increased potential for carriage and dissemination of pathogens within human populations and facilitated emergence of resistant human pathogens.
____
The environmental impact of inappropriate antimicrobial use in animals:
Another area of human health concern is the long-term effect of antibiotic residues in the environment. Although human antimicrobial usage may be the primary source for aquatic and terrestrial antibiotic contamination, antibiotic applications in livestock, poultry and aquaculture also contribute significantly to this growing problem. A varying proportion of administered antibiotics may remain active in excreted biological matter (generally feces or urine) after passing through the animal. Along with antimicrobials used for humans, the livestock, poultry and aquaculture sectors are important contributors to aquatic and terrestrial contamination with antibiotics. Antibiotics and their metabolites (degradation products) reach the environment via the application of antibiotic-laden manure or slurry on agricultural lands, or direct deposition of manure by grazing animals. This can be followed by surface run-off, driftage or leaching into deeper layers of the earth. A proportion of the antibiotics that reach the environment will remain biologically active. Low subtherapeutic concentrations of antibiotics that accumulate over time may have profound effects on some ecosystems. Environmental antibiotic concentrations may exert selective pressure on environmental bacteria and may also foster the transfer of resistance genes, helping create the “resistome” mixing pot of genetic AMR traits.
_
Antibiotics in the Environment:
_
Environmental antibiotic pollution encourages the transfer of resistance genes to human commensal and pathogenic bacteria. In particular, waste water treatment plants serving antibiotic manufacturing facilities have been implicated in the transfer of resistance genes into human microbiota and pose a serious threat to antibiotic effectiveness given the size of India’s pharmaceutical sector. Waters polluted by the ordure of pigs, poultry, or cattle represent a reservoir of antibiotic resistance genes, both known and potentially novel. These resistance genes can be spread among different bacterial species by bacteriophage, bacteria-infecting viruses. Antibiotic resistance genes have been found in bacteria in drinking water and sewage, far the hospitals the usually haunt. Bacteria carrying a gene that confers resistance to a major class of antibiotics have shown up in samples of drinking water and sewage seepage from New Delhi, researchers report in The Lancet Infectious Diseases. Research data suggest that possibly about 10% of the people in New Delhi carry NDM-1 in normal flora in their guts. If those native gut flora end up passing on resistance to harmful strains of E. coli, for example, that could lead to infections, such as cystitis, with virtually no drugs that can treat it. Whenever an antibiotic is used/overused/ misused, resistance develops. A mix of bacteria enters the sewage, contaminates drinking water and enters the gut of a healthy individual, making him/her resistant to those bugs. This resistance moves from one level to another.
__
Wildlife and antimicrobial resistance:
A growing number of studies — in crows, elephant seals, voles and other wild animals — are raising big questions about where wildlife fits into the increasing threat of antimicrobial resistance. Genes for resistance are showing up in microbes flourishing in the guts and other parts of wild animals. How those genes get there and where they might go now needs serious attention. So far, scientists have not described a clear-cut case of genes for antimicrobial resistance traveling from wildlife microbial flora back to humans, but that scenario is “very biologically logical,” says Barry McMahon of University College Dublin. McMahon, who has examined gulls for antimicrobial resistance genes, says overlooking wildlife and environmental factors leaves a big gap in understanding resistance. So does Kathleen Alexander of Virginia Tech in Blacksburg. Monitoring what’s circulating in wild animals might serve as early warnings for what’s ahead. Focusing solely on hospitals, she says, is “monitoring the barn after the horse has left.” Genes for resistance can readily spread as bacteria multiply and carry their toolkits with them. And bacteria are “promiscuous”. They commingle genes with their own kind or with fairly strange strangers, widely distributing resistance genes. In this loose networking, a benign bacterium can pass along resistance genes to a pathogen, especially as resistance turns up in microbes in a wide diversity of animals. Reports come from places with minimal local people or livestock to pass along resistance picked up during medical treatment. Among 97 birds checked in the Arctic (Siberia, Alaska and Greenland), researchers in 2008 reported Escherichia coli bacteria resistant to 14 of the 17 antibiotics they tested. Admittedly birds fly, but monkeys don’t. In the Uxpanapa forests of Mexico, however, howler monkeys had E. coli resistance to ciprofloxacin, a synthetic antibiotic. That suggests some connection, however roundabout, between human medicine and faraway monkeys. Maybe the answer is birds flying in and roosting in trees. But for any resistance transfer involving wildlife, the forensic trail isn’t well understood. To date, researchers have only circumstantial evidence, much of it involving runoff from human wastes. A 2008 study of stranded northern elephant seals along the California coast, for instance, found that the nearer the animals were to outflow of freshwater from land, the more likely they were to test positive for antimicrobial-resistant E. coli.
________
Genetically modified organisms (GMO) and AMR:
Antibiotic resistance in genetically modified crops:
Antibiotic-resistance genes are used as “markers” in genetically modified crops. The genes are inserted into the plant in early stages of development to in order to detect specific genes of interest. e.g. herbicide-resistant genes or insecticidal toxin genes. Approximately 70% of all transgenic plants have been produced using the neomycin phosphotransferase II (nptII) gene from Escherichia coli for good reason: it works, especially in most dicot species. The antibiotic-resistance genes have no further role to play, but they are not removed from the final product. This practice has met with criticism because of the potential that the antibiotic-resistance genes could be acquired by microbes in the environment. In some cases these marker genes confer resistance to front-line antibiotics such as the beta-lactams and aminoglycosides. The movement of a transgene from plant to microbe could pose a significant risk, especially if an antibiotic resistance gene, originally from a bacterium, could be transferred to a pathogenic bacterium, causing new antibiotic resistance problems for human health. These concerns have prompted regulators and companies alike to look askance at the use of antibiotic resistance genes in transgenic plants.
_
GMOs’ Antibiotic Resistance and Food Safety:
Unlike safety evaluations for drugs, there are no human clinical trials of GM foods. The only published human feeding experiment revealed that the genetic material inserted into GM soy transfers into bacteria living inside our intestines and continues to function. This means that long after we stop eating GM foods, we may still have their GM proteins produced continuously inside us. This could mean:
1. If the antibiotic gene inserted into most GM crops were to transfer, it could create super diseases, resistant to antibiotics.
2. If the gene that creates Bt-toxin in GM corn were to transfer, it might turn our intestinal bacteria into living pesticide factories.
Although no studies have evaluated if antibiotic or Bt-toxin genes transfer, that is one of the key problems. The safety assessments are too superficial to even identify most of the potential dangers from GMOs.
________
________
The figure below shows dissemination of antibiotics and antibiotic resistance within agriculture, community, hospital, wastewater treatment, and associated environments.
______
______
The figure below shows interactions of various influences on development of AMR:
There are four main contributors to the development of bacterial antibiotic resistance are: the pharmaceutical industry, the agriculture and animal husbandry industry, patients, and healthcare providers.
______
_______
Laboratory Diagnosis of AMR:
_
Detecting antimicrobial resistance:
Antimicrobial susceptibility testing methods are in vitro procedures used to detect antimicrobial resistance in individual bacterial isolates. Because these laboratory detection methods can determine resistance or susceptibility of an isolate against an array of possible therapeutic candidates, antimicrobial susceptibility testing results can be a useful clinical guideline in selecting the best antibiotic treatment option for each particular patient. These same methods can also be used for monitoring the emergence and spread of resistant microorganisms in the population. Clinical Breakpoints are threshold values established for each pathogen-antibiotic (i.e., bug-drug) combination indicating at what level of antibiotic the isolate should be considered to be sensitive, intermediate or resistant. The interpretative criteria for these are based on extensive studies that correlate laboratory resistance data with serum achievable levels for each antimicrobial agent and a history of successful and unsuccessful therapeutic outcomes. Standard conditions for these assays have been established based on extensive batteries of laboratory testing. Guidelines and recommendations for these are continuously updated by certain organizations worldwide. Although veterinary laboratories originally based interpretations on standards established using human pathogens, it became apparent by the early 1980s that such an approach did not reliably predict clinical outcomes when applied to veterinary practice. Subsequently, groups within organizations that set standards were created for the purpose of developing veterinary-specific standards.
_
Lab approaches and strategies:
Points to consider when deciding whether or not to conduct antimicrobial susceptibility testing should include:
•clinical relevance of the isolate
•purity of the isolate
•logical panel of antimicrobial agents to be tested (i.e., do not include antibiotics to which the isolate is known to have intrinsic resistance)
•availability of test methodology, resources, and trained personnel
•standardization of testing
•valid interpretation of results
•cost efficiency
•effective means to communicate results and interpretation to end-users
_
Most often, interpretation is reduced to whether the isolate is classified as susceptible, intermediately susceptible, or resistant to a particular antibiotic. It should, however, be remembered that these in vitro procedures are only approximations of in vivo conditions which can be very different depending on the nature of the drug, the nature of the host and the conditions surrounding the interaction between the antibiotic and the target pathogen. In Pakistan, laboratory data indicate that 78% of pneumococci are resistant to cotrimoxazole, yet the clinical treatment failure rate is only 15%. The reasons for this discrepancy are unknown, but the issue is important because alternative drugs are more expensive. Standardization of laboratory methods and appropriate surveillance methods are essential. One critical aspect is following standardized procedures that can generate reproducible results, i.e. quality control.
Aspects of quality control include:
•standardized bacterial inoculum size
•culture conditions (growth medium, pH, cation concentration, blood and serum supplements and thymidine content)
•incubation conditions (atmosphere, temperature, duration)
•concentration of antimicrobials for testing.
Because of the required culture time, antimicrobial susceptibility testing may take several days, which is not ideal particularly in critical clinical cases demanding urgency. Often, practitioners may utilize locally established antibiograms as guideline for therapy. An antibiogram is a compiled susceptibility report or table of commonly isolated organisms in a particular hospital, farm, or geographic area, which can serve as a useful guideline in therapy before actual culture and susceptibility data becomes available for reference.
__
Test Methods in Detecting Antimicrobial Resistance:
There are several antimicrobial susceptibility testing methods available today, and each one has their respective advantages and disadvantages. They all have one and the same goal, which is to provide a reliable prediction of whether an infection caused by a bacterial isolate will respond therapeutically to a particular antibiotic treatment. This data may be utilized as guidelines for chemotherapy, or at the population level as indicators of emergence and spread of resistance based on passive or active surveillance. Some examples of antibiotic sensitivity testing methods are:
•Dilution method (broth and agar dilution method)
•Disk-diffusion method
•E-test
•Automated methods
•Mechanism-specific tests such as beta-lactamase detection test and chromogenic cephalosporin test
•Genotypic methods such as PCR and DNA hybridization methods
Selection of the appropriate method will depend on the intended degree of accuracy, convenience, urgency, availability of resources, availability of technical expertise and cost. Interpretation should be based on human medical standards.
_
1. Dilution Methods:
The Broth dilution method involves subjecting the isolate to a series of concentrations of antimicrobial agents in a broth environment. Microdilution testing uses about 0.05 to 0.1 ml total broth volume and can be conveniently performed in a microtiter format. Macrodilution testing uses broth volumes at about 1.0 ml in standard test tubes. For both of these broth dilution methods, the lowest concentration at which the isolate is completely inhibited (as evidenced by the absence of visible bacterial growth) is recorded as the minimal inhibitory concentration or MIC. The MIC is thus the minimum concentration of the antibiotic that will inhibit this particular isolate. The test is only valid if the positive control shows growth and the negative control shows no growth. A procedure similar to broth dilution is agar dilution. Agar dilution method follows the principle of establishing the lowest concentration of the serially diluted antibiotic concentration at which bacterial growth is still inhibited. On agar plate, a bacterial isolate is tested for resistance to each of twelve different antibiotics. The clear zones around each disc are the zones of inhibition that indicate the extent of the test organism’s inability to survive in the presence of the test antibiotic. Presence of zone of inhibition is not automatically interpreted as susceptibility to the antibiotic; the zone width has to be measured and compared against a reference standard which contains measurement ranges and their equivalent qualitative categories of susceptible, intermediately susceptible or resistant.
For example, one E.coli isolate has a zone of inhibition of 10.1mm around ampicillin (AM); since the zone diameter interpretation chart is as follows:
Resistant: 13mm or less
Intermediate: 14-16 mm
Susceptible: 17 mm or more
This particular E.coli isolate is read as resistant to ampicillin.
_
2. Disk Diffusion Method:
Because of convenience, efficiency and cost, the disk diffusion method is probably the most widely used method for determining antimicrobial resistance in hospitals and clinics. A growth medium is first evenly seeded throughout the plate with the isolate of interest that has been diluted at a standard concentration. Commercially prepared disks, each of which are pre-impregnated with a standard concentration of a particular antibiotic, are then evenly dispensed and lightly pressed onto the agar surface. The test antibiotic immediately begins to diffuse outward from the disks, creating a gradient of antibiotic concentration in the agar such that the highest concentration is found close to the disk with decreasing concentrations further away from the disk. After an overnight incubation, the bacterial growth around each disc is observed. If the test isolate is susceptible to a particular antibiotic, a clear area of “no growth” will be observed around that particular disk. The zone around an antibiotic disk that has no growth is referred to as the zone of inhibition since this approximates the minimum antibiotic concentration sufficient to prevent growth of the test isolate. This zone is then measured in mm and compared to a standard interpretation chart used to categorize the isolate as susceptible, intermediately susceptible or resistant.
Antibacterial Sensitivity Tests (AST) = drug sensitivity test (DST):
Some antibacterial medicines are more active against gram-positive than gram-negative bacteria and others are the reverse. This gives a guide to the choice of medicine to be used. Medicine sensitivity tests carried out in laboratories give a better guide against specific infections, but this is not a perfect one. Some bacteria which are sensitive in laboratory tests are not sensitive in diseased individuals. They may be multiplying in sites where the medicine cannot reach them, or the antibiotic is not reaching them in high enough concentrations. In its simplest and commonest form the test is carried out by growing the bacteria in a growth medium and then suspending them in a saline solution. A thin film of this is spread over the surface of a culture plate and left to dry. Discs of cardboard impregnated with different antibiotics are placed on the surface and the plate is then incubated. The medicine diffuses out into the growth medium radially. If the organism is killed by the antibiotic there is a clear zone of no growth around the disc. If the organism is resistant to the medicine it grows right up to the disc. Such tests usually take 12 – 24 hrs. Some bacteria however take weeks to grow.
_
As seen in the figure below, Bacteria are streaked on dishes with white antibiotic impregnated disks. Clear rings, such as those on the left, show that bacteria have not grown — indicating that the bacteria are not resistant. Those on the right are fully susceptible to only three of the seven antibiotics tested.
_
Antimicrobial susceptibility testing (AST) methods are in vitro procedures used to detect antimicrobial resistance in individual bacterial isolates. Because these laboratory detection methods can determine resistance or susceptibility of an isolate against an array of possible therapeutic candidates, AST results can be a useful guideline in selecting the best antibiotic treatment option for each particular patient. Examples of AST methods are: broth (and agar) dilution methods, disk-diffusion test, E-test, automated detection using various commercially available detection kits, mechanism-specific methods such as those which detect specific enzymes that bring about resistance, and by applying genotypic methods which detect antibiotic resistance genes.
_
3. E-Test:
E-test (AB Biodisk, Solna, Sweden) is a commercially available test that utilizes a plastic test strip impregnated with a gradually decreasing concentration of a particular antibiotic. The strip also displays a numerical scale that corresponds to the antibiotic concentration contained therein. This method provides for a convenient quantitative test of antibiotic resistance of a clinical isolate. However, a separate strip is needed for each antibiotic, and therefore the cost of this method can be high.
_
4. Automated Antimicrobial Susceptibility Testing Systems:
Several commercial systems have been developed that provide conveniently prepared and formatted microdilution panels as well as instrumentation and automated reading of plates. These methods are intended to reduce technical errors and lengthy preparation times. Most automated antimicrobial susceptibility testing systems provide automated inoculation, reading and interpretation. These systems have the advantage of being rapid (some results can be generated within hours) and convenient, but one major limitation for most laboratories is the cost entailed in initial purchase, operation and maintenance of the machinery. Some examples of these include: Vitek System (bioMerieux, France), Walk-Away System (Dade International, Sacramento, Calif.), Sensititre ARIS (Trek Diagnostic Systems, East Grinstead, UK), Avantage Test System (Abbott Laboratories, Irving, Texas), Micronaut (Merlin, Bornheim-Hesel, Germany), Phoenix (BD Biosciences, Maryland) and many more.
_
5. Mechanism-Specific Tests:
Resistance may also be established through tests that directly detect the presence of a particular resistance mechanism. For example, beta lactamase detection can be accomplished using an assay such as the chromogenic cephalosporinase test (Cefinase disk by BD Microbiology Systems, Cockeysville, MD and BBL DrySlide Nitrocefin, Becton Dickinson, Sparks, MD) and detection for chloramphenicol modifying enzyme chloramphenicol acetyltransferase (CAT) may utilize commercial colorimetric assays such as a CAT reagent kit (Remel, Lenexa, Kansas). Nitrocefin is a chromogenic cephalosporin substrate routinely used to detect the presence of beta-lactamase enzymes produced by various microbes. Other methods for beta-lactamase detection exist including PCR; however, nitrocefin allows for rapid beta-lactamase detection using few materials and inexpensive equipment.
_
6. Genotypic Methods:
Since resistance traits are genetically encoded, we can sometimes test for the specific genes that confer antibiotic resistance. Genotypic methods are also known as molecular assays. However, although nucleic acid-based detections systems are generally rapid and sensitive, it is important to remember that the presence of a resistance gene does not necessarily equate to treatment failure, because resistance is also dependent on the mode and level of expression of these genes. Some of the most common molecular techniques utilized for antimicrobial resistance detection are as follows
a) Polymerase chain reaction (PCR) is one of the most commonly used molecular techniques for detecting certain DNA sequences of interest. This involves several cycles of denaturation of sample DNA, annealing of specific primers to the target sequence (if present), and the extension of this sequence as facilitated by a thermostable polymerase leading to replication of a duplicate DNA sequence, in an exponential manner, to a point which will be visibly detectable by gel electrophoresis with the aid of a DNA-intercalating chemical which fluoresces under UV light.
b) DNA hybridization: This is based on the fact that the DNA pyrimidines (cytosine and thymidine) specifically pair up with purines (guanine and adenine; or uracil for RNA). Therefore, a labelled probe with a known specific sequence can pair up with opened or denatured DNA from the test sample, as long as their sequences complement each other. If this “hybridization” occurs, the probe labels this with a detectable radioactive isotope, antigenic substrate, enzyme or chemiluminescent compound. Whereas if no target sequence is present or the isolate does not have the specific gene of interest, no attachment of probes will occur, and therefore no signals will be detected.
c) Modifications of PCR and DNA hybridization: With these basic principles, several modifications have been introduced which further improve the sensitivity and specificity of these standard procedures. Examples of such development were the use of 5’-fluorescence-labeled oligonucleotides, the development of molecular beacons, development of DNA arrays and DNA chips, among many others.
_
Pros and cons of genotypic methods:
DNA sequencing has several advantages. First, by definition, sequencing DNA is the most comprehensive technique for determining genotypes. Second, it can be applied to determine alleles of resistance loci and clonal backgrounds, using the method known as multi-locus sequence typing which typically involves sequencing several hundred base pairs of DNA at multiple loci scattered around the genome. Third, it is portable among facilities because the data are informationally simple (four nucleotides in a unitary string). Fourth, sequence data can be readily adapted for population genetic analyses that can be used for the evolution of resistance clones. Finally, sequence data also lend themselves to molecular analyses that could identify regions of resistance loci that affect function. DNA sequencing has two major disadvantages. The first is cost. Most sequencing facilities can perform MLST at a cost of roughly $40–$80 US per isolate, so large numbers of isolates often exceed the resources of many investigators. The other disadvantage is a lack of temporal resolution. When two isolates differ by only one nucleotide out of approximately 3000 examined, it is difficult to determine when that divergence occurred: it may have occurred yesterday, or, in the case of E. coli, roughly 200 years ago.
____
Genotype vs. phenotype testing:
There has long been a debate over whether the microbial genotype or phenotype is a better determinant of clinical response when looking at antimicrobial resistance testing. The genotype for antimicrobial susceptibility testing identifies genes within the microbe that have the potential to cause resistance. However it does not provide information on whether the gene is “switched on”. The phenotype is how the microbe responds to the antimicrobial in the laboratory setting, but would not pick up resistance potential, and in particular resistance genes that may code for “inducible” resistance. There has been a trend in recent years for a move towards calling results from genotypic testing the “gold standard”. This is probably due to two factors:
1. Increased range of molecular assays for resistance determinants available.
2. Fear of not knowing the genotype and thus missing the potential for resistance.
However genotypic resistance testing may well be over-calling anti-microbial resistance in some cases, and thus pushing patients unnecessarily towards broader spectrum antibiotics, thus starting a vicious circle of resistance. This is not something however, that is often brought up in the anti-microbial stewardship committee setting. On the other hand, genotypic testing is generally rapid and sensitive resulting in early detection of AMR allowing clinicians to change drugs quickly before bacterial/viral load increases and patient becomes severely ill.
____
____
Laboratory Diagnosis of MDR-TB AND XDR-TB:
Drug-resistant TB often goes undetected and untreated in many countries. With the exception of a few developed countries, most national TB programs worldwide do not routinely provide diagnostic services based on culture and DST. The laboratory is an essential component in TB control programs, and broader access to DST is a priority for most countries. Early choice of appropriate treatment is an essential determinant of favourable outcome, and rapid determination of drug resistance can allow a customized approach to treatment early in the course of the disease and can potentially reduce morbidity, mortality and infectiousness. The diagnosis of MDR-TB and XDR-TB is hampered by the absence of effective and affordable rapid diagnostic techniques for drug sensitivity. Several approaches, phenotypic and molecular, have been explored to develop rapid, reliable and accurate methods for the rapid detection of drug resistance in M tuberculosis. These methods should also be evaluated and applied in high-incidence areas.
1. CONVENTIONAL CULTURE-BASED METHODS:
Using standardized DST procedures with conventional methods, eight to 12 weeks are required to identify drug-resistant microorganisms on solid media (i.e., Lowenstein-Jensen medium). In general, such methods assess inhibition of M tuberculosis growth in the presence of antibiotics to distinguish between susceptible and resistant strains. The proportion method allows precise determination of the proportion of resistant mutants to a certain drug; the resistance ratio method compares the resistance of an unknown strain with that of a standard laboratory strain. While relatively inexpensive and undemanding of sophisticated equipment, results usually take weeks and this is challenging; inappropriate choice of treatment regimen may result in death within weeks of initiation, such as in the case of XDR-TB (especially in HIV-infected patients). In addition, delayed identification of drug resistance results in inadequate treatment, which may generate additional drug resistance and continued transmission in the community.
2. LIQUID CULTURE-BASED METHODS:
Automated liquid culture systems are more sensitive than solid media cultures, and they significantly reduce turnaround time. However, even with liquid cultures, two to four weeks are still needed to obtain results, and their substantially higher cost is an issue for resource-limited countries. The BACTEC 460 TB radiometric system (Becton Dickinson, USA) was considered to be a major advancement when it was introduced, but has been replaced by the Mycobacteria Growth Indicator Tube system (Becton Dickinson, USA). Several published studies have shown the excellent performance of the Mycobacteria Growth Indicator Tube system for the rapid detection of resistance to first- and second-line anti-TB drugs. Detection of drug resistance can be accomplished in days rather than weeks, although still constrained by high cost (equipment and consumables).
3. NOVEL, RAPID PHENOTYPIC METHODS:
Among novel, rapid phenotypic methods, the microcolony method is relatively low cost. It has been adapted for the rapid detection of drug resistance directly from sputum samples, and has been shown in early studies to be accurate for the detection of MDR-TB compared with the reference proportion method, with results available in one week. Newly developed phenotypic tests such as TK Medium (Salubris Inc, USA), microscopic-observation drug-susceptibility assay (MODS) and FASTPlaque-Response bateriophage assay (Biotec Laboratories Ltd, UK) are usually cheaper but not always simple to perform, with some requiring high standards of biosafety and quality control.TK Medium is a novel colorimetric system that indicates growth of mycobacteria by changing the colour of the growth medium. Metabolic activity of growing mycobacteria changes the colour of the culture medium, and this allows for an early positive identification before bacterial colonies appear. TK Medium also permits susceptibility testing for drug resistance, and can allow for differentiation between M tuberculosis and nontuberculous mycobacteria. Unfortunately, there is insufficient published evidence on the field performance of this test in developing countries. The MODS assay is based on the observation of the characteristic cord formation of M tuberculosis that is visualized microscopically in liquid medium with the use of an inverted microscope. MODS uses simple light microscopy to detect early growth of M tuberculosis as ‘strings and tangles’ of bacterial cells in the broth medium with or without antimicrobial drugs (for DST) (12). The agreement between MODS and the reference standard for drug susceptibility testing is 97% for INH, 100% for RIF, and 99% for INH and RIF combined (MDR). Lower values of agreement were obtained for ethambutol (95%) and streptomycin (92%). One minor disadvantage of MODS is the requirement for an inverted microscope for observation of the mycobacterial growth. FASTPlaque-Response is a phage amplification-based test, and has been developed for direct use on sputum specimens. Drug resistance is diagnosed when M tuberculosis is detected in samples that contain the drug (i.e., RIF). A recent meta-analysis of the accuracy of phage-based methods for detecting RIF resistance in M tuberculosis concluded that these assays performed on M tuberculosis culture isolates have high sensitivity, but variable and slightly lower specificity. Not enough evidence is available on the accuracy of these assays when performed directly on sputum samples. Safety and quality control issues related to the use of this technique should also be addressed carefully. Several colorimetric methods have also been proposed in the past few years for the rapid detection of drug resistance in M tuberculosis. A recent systematic review and meta-analysis of colorimetric redox indicator methods found evidence of high sensitivity and high specificity for the rapid detection of MDR-TB. Colorimetric methods represent a good alternative for the rapid detection of drug resistance in laboratories with limited resources. However, these tests cannot be directly used on clinical specimens.
4. NOVEL, RAPID MOLECULAR METHODS:
The identification of specific mutations responsible for drug resistance has facilitated the development of novel, rapid molecular tools for DST. The detection of RIF resistance is traditionally used as a predictor of MDR-TB – its positive predictive value is a function of the sensitivity and specificity of RIF resistance testing and the prevalence of MDR and non-MDR RIF resistance, which is highest among previously treated cases in settings with high MDR prevalence and low non-MDR RIF resistance. Molecular tools are based on nucleic acid amplification in conjunction with electrophoresis, sequencing or hybridization. Although most of the techniques were initially developed to detect drug resistance in TB complex isolates, they are being evaluated for direct detection of TB complex isolates and identification of alleles related to drug resistance in clinical specimens (such as sputum). Their potential advantage is that there is no need for growth of the organism and DST results can be determined in days rather than weeks; research suggests that they can be highly reliable.
Molecular Assays:
PCR based technologies using various modifications are employed to detect the presence of putative resistance genes (rpoβ for rifampicin, katG and inhA for INH etc.).
Line probe assay:
Reverse hybridization-based assays, referred to as line probe assays, represent a useful tool for their superior cost-effectiveness. These tests are based on the hybridization of specific probes for wild-type and mutated sequences of genes involved in drug resistance, and they show high specificity and medium/high sensitivity. Line Probe Assays (LPA) is based on in-situ hybridization on nitrocellulose strips of specific genetic targets for resistance genes. The newer Genotype® MTBDRplus detects additional mutations in the katG gene and also in the inhA promoter region for INH resistance, leading to a higher sensitivity. It was also found pooled sensitivity and specificity of 98% and 99% respectively, for RIF but for INH, sensitivity was 84%, though specificity was 100%. These are for smear positive pulmonary TB cases. Commercially available line probe assays include the INNO-LiPA Rif. TB kit (Innogenetics, Belgium) and the GenoType MTBDR assay (Hain Lifescience, Germany). A recent meta-analysis summarized the results obtained for the INNO-LiPA Rif. TB test, and showed that this line probe assay has high sensitivity and specificity when culture isolates are used. The majority of studies had sensitivities of 95% or greater, and nearly all were 100% specific. The results, however, are less accurate when the test is directly applied to clinical specimens (i.e., sputum). There is a paucity of data on the application of this test directly to clinical specimens.
Cepheid GeneXpert:
It is a completely closed automated system using real-time PCR having a sensitivity of 70 – 90% even for smear negative cases. It can detect the presence of RIF resistance. The advantage of this system is that the infrastructure and manpower requirements are minimal, making it an ideal tool for achieving same day diagnosis and DST on the same day at the periphery, so that the patients can be diagnosed efficiently and put quickly on an effective treatment regimen.
Although MDR-TB is defined as resistance to at least INH and Rifampicin, the key determinant for treatment failure is Rifampicin resistance. Detection of Rifampicin resistance has thus been proposed as a proxy for MDR-TB diagnosis, as well as for epidemiological monitoring. DST for Rifampicin, by conventional methods based on growth as well as by newer genetic techniques, is generally considered the most reliable.
______
______
Tests for malaria Drug Resistance:
CDC recommends that all cases of malaria diagnosed in the United States should be evaluated for drug resistance. Specimens from all cases diagnosed in the United States should be sent to the CDC Malaria Laboratory for this testing. All specimens submitted for drug resistance testing will first undergo molecular testing by polymerase chain reaction (PCR) for species confirmation. Depending on the species, the age of the specimen, and the previous receipt of antimalaria medicines, different testing methods may be employed to assess for the presence of drug resistance. Laboratory methods for testing for malaria drug resistance include in vitro tests, molecular characterization, determination of drug levels, and animal models.
1. In vitro tests:
In these tests, blood samples from malaria patients are cultured and if found to be viable, the malaria parasites are exposed to different concentrations of antimalarial drugs in the laboratory. Culturing malaria parasites will be attempted only for pre-treatment specimens that arrive within 72 hours of collection.
2. Molecular characterization:
For some drugs (chloroquine, SP and similar drugs, atovaquone), molecular markers have been identified that confer resistance. Molecular techniques, such as PCR or gene sequencing can identify these markers in blood taken from malaria-infected patients.
3. Drug levels:
When individuals who are presumably taking malaria prophylaxis develop malaria, blood samples can be collected and analyzed for the presence of the drug. The drug is extracted from the blood and the concentration is determined by high-performance liquid chromatographic methods. Determining the drug level in the blood can help distinguish between drug non-adherence and drug resistance.
_______
_______
Drug Susceptibility Testing for HIV:
Drug susceptibility testing involves culturing a fixed inoculum of HIV-1 in the presence of serial dilutions of an inhibitory drug. The concentrations of drug required to inhibit virus replication by 50% (IC50) or 90% (IC90) are the most commonly used measures of drug susceptibility. Drug susceptibility results depend on the inoculum size of virus tested, the cells used for virus replication, and the means of assessing virus replication. Drug susceptibility assays are not designed to determine the exact amount of drug required to inhibit virus replication in vivo but rather to identify differences in the drug concentration required to inhibit a fixed inoculum of a virus relative to the concentrations required to inhibit wild-type viruses. Virus susceptibility to a drug can be characterized by the range in susceptibility obtained testing wild-type virus isolates (wild-type susceptibility range) and the range in susceptibility obtained testing resistant virus isolates (dynamic susceptibility range).
_
Generally speaking, there are two types of drug-resistance tests available to HIV-positive patients: genotypic and phenotypic assays. Because genotypic testing provides results in one to two weeks—compared to the two-, three- or four-week turnaround associated with phenotypic testing—it is the preferred choice for patients who have yet to start treatment. For people whose HIV therapy has stopped working, DHHS Treatment guidelines recommend that resistance testing also be used to confirm treatment failure and to help select a new regimen. When a first or second regimen has failed, genotypic testing is preferred. Phenotypic testing, say the guidelines, should be used when a person has more extensive drug resistance. Among patients who have used multiple drugs in the past, interpreting the results of both tests together may be most useful. One company, Monogram Biosciences, will phenotype and genotype a blood sample and provide the results of both tests on the same lab report—called PhenoSense GT. In general, both tests work best when a person has a viral load of 500 or more, and preferably at least 1,000.
_
Genotypic resistance testing for HIV:
These tests examine the actual genetic structure—or genotype—of HIV taken from a patient (a standard blood sample is all that is required). The HIV is examined for the presence of specific genetic mutations that are known to cause resistance to certain drugs. An example: Researchers know that the NRTIs Epivir and Emtriva are not effective against forms of HIV that contain the mutation “M184V” in its reverse transcriptase gene. If a genotypic resistance test discovers a mutation at position M184V, chances are that the person’s HIV is resistant to Epivir and Emtriva and is not likely to respond to either of these drugs. For many drugs, including the protease inhibitors and other NRTIs, complex patterns of mutations are required for resistance to occur. In this way, interpreting the results of genotypic testing can be tricky, given that different mutations—and different combinations of mutations, especially in patients with a lot of treatment experience—can mean different things. However, as knowledge of mutations and their different patterns has grown considerably over the past several years, laboratories are able to provide accurate and useful information to physicians. Examples of available genotypic tests include: Bayer Health Diagnostics’ HIV-1 TrueGene, Celera Diagnostics/Abbott Laboratories’ ViroSeq, LabCorp’s GenoSure (Plus) and Monogram Biosciences’ GeneSeq.
_
_
Phenotypic resistance testing for HIV:
Unlike genotypic testing, which looks for particular genetic mutations that cause drug resistance, phenotypic testing directly measures the behavior—or phenotype—of a patient’s HIV in response to particular antiretrovirals. Because of the way phenotypic tests work and the results they provide, many experts believe that these tests are more comprehensive and trustworthy than genotypic tests, especially when testing samples from patients who have tried and failed a number of HIV drugs in the past. Using the simplest terms, phenotypic testing is performed by placing samples of a patient’s HIV in test tubes with each HIV drug to observe how the virus reacts. The ability of the virus to grow (or not grow) in the presence of each drug is evaluated. The virus is exposed to varying strengths, or concentrations, of each drug. The ability of the patient’s virus to grow in the presence of the drugs is compared with some wild-type virus that is known to be 100 percent susceptible to all HIV drugs. The comparison between the patient’s virus and the wild-type virus provides the phenotyping results. These results tell doctors how much of a particular drug is needed to reduce HIV replication. In other words, the laboratory conducting a phenotypic test is trying to determine the amount, or concentration, of drug needed to stop HIV from reproducing. For example, if four times as much of the NRTI abacavir is needed to control HIV replication, the virus is said to have “fourfold resistance” to the drug. If seven times as much is needed, the virus is sevenfold resistant to the drug. After several years of extensive research, these companies have developed “clinical cutoffs”—an important component of phenotypic testing that allows for much easier interpretation of fold changes as they relate to the sensitivity of HIV to many of the available medications. Each HIV drug has different clinical cutoffs, which can be confusing. To help make sense of these cutoffs and to make it easier for health care providers to interpret the results, laboratories conducting these tests provide detailed reports for every test conducted. There are two “conventional” phenotypic tests available: Monogram Bioscience’s PhenoSense assay and Virco Lab’s Antivirogram. Both tests evaluate the fold changes for all of the available NRTIs, Protease Inhibitors, and NNRTIs. Monogram Biosciences has a separate phenotypic assay, called PhenoSense Entry, which tests HIV’s sensitivity to the entry inhibitor Fuzeon. Another test is Virco Lab’s vircoTYPE HIV-1 assay. This is actually a “predictive” phenotypic test, using genotypic testing results to figure out what the virus’s phenotype is, without actually performing a phenotypic test. To do this, labs use genotyping testing to determine if an HIV sample has mutations known to cause drug resistance. Once the genotype has been determined, the laboratory searches a database maintained by Virco containing the genotypes of several thousand HIV samples collected from other patients. It then retrieves the phenotypes—the fold changes—that correspond to these samples, averages the information together and predicts the drugs that the current sample will be more or less sensitive to. It is important to note that Monogram Biosciences and Virco calculate their cutoffs differently. As a result, the cutoffs determined for one company’s test (e.g., PhenoSense) do not apply to the cutoffs determined for the other company’s test (e.g., vircoTYPE).
______
______
New Technologies to detect Antimicrobial Resistance more quickly:
Several companies are exploiting new sensitive technologies, such as magnetic resonance technology, fluorescence in situ hybridization, and transcriptional profiling to enable pathogen detection directly from a blood sample and eliminate the need for a culture step. In addition, distinguishing between colonization and infection at nonsterile sites presents a distinct technical challenge. For example, in respiratory tract infections, detected organisms may not be the cause of the patient’s symptoms. To circumvent these issues, researchers are developing tests based on host response expression signatures, which could help distinguish colonization from infection and bacterial from viral infections. Biomarkers, such as procalcitonin, are used in some countries as surrogates of infection to support microbial diagnosis, and they are also being explored as tools to help guide initiation of empirical therapy. Discerning whether an infection is viral or bacterial can be challenging. This uncertainty accounts for much of the overuse and over-prescribing of broad spectrum antibiotics. In many cases, illnesses such as respiratory infections, skin infections, and urinary tract infections (UTI) are viral or could be treated by antibiotics that are less likely to cause resistance. Current traditional culture methods can take up to three days to identify bacteria and test for antimicrobial resistance in a urine sample. Genome sequencing will be increasingly used in the clinical setting to tailor antimicrobial prescribing and inform infection control outbreaks. A recent technological innovation that could reduce the delay between pathogen sampling and data generation is single molecule sequencing. An example of this technology, which is undergoing evaluation through an early access programme, is the Oxford Nanopore MinION. For this study, the researchers used human cells removed from urine samples. Bacteria was recovered and sequenced by MinION. The results were then compared with standard culture and antibiotic susceptibility testing results. In order to detect sub-variant chromosomal genes within a cluster and distinguish them between TEM-1 and TEM52, MinION must be able to identify mutations in relation to antimicrobial resistance. The researchers concluded that although MinION detects the presence of acquired resistance genes, in order for the results to accurately predict mutations in chromosomal genes, improvements to the technology are necessary.
____
Creating and predicting resistance profiles:
Baym and his colleagues envision a scenario in which hospitals can rapidly sequence the genome of a patient’s infection and use that information to identify which drugs a bacteria is resistant to and how that resistance works. Such detailed information would allow doctors to choose the most effective drug combination and even reconstruct a pathogen’s evolutionary history. That scenario may not be too distant. An Oxford University-based project called Mykrobe is planning a trial of a program called Predictor, which will analyze bacterial genomes for genes related to resistance and then tell doctors which drugs the pathogen is resistant to. So far, the Mykrobe team has designed a version for MRSA and one for tuberculosis. Of course, tools like Predictor still need genome data to work with, which means the bacteria’s DNA has to be sequenced quickly enough to be useful for treatment. Making it practical for the average hospital to sequence bacterial genes in real time is a significant challenge. The technology exists, but it’s not fast enough or affordable enough for most hospitals to use as a clinical tool. Meanwhile, Baym and his colleagues have even more ambitious ideas about the future of the arms race against resistant bacteria. They expect doctors to someday be able to get ahead of bacteria by predicting how a strain might evolve and then moving preemptively to counter it. “For example, if a pathogen’s genome is a few mutations away from resistance, we might predict that, while it is not resistant, it could become resistant if certain drugs are used,” they wrote. For a given drug, there are only a limited number of ways that bacteria can resist. That means it could be possible to at least predict the probability of mutations that lead to each one; it’s easy to imagine mathematical approaches lay the groundwork for such techniques. However, any kind of prediction would require more detailed knowledge about the genetics behind antibiotic resistance. That’s something that scientists around the world are still working on. Ultimately, just as bacteria can evolve several different ways to resist antibiotics, doctors will need multiple strategies to thwart resistance. There is, as Baym and his colleagues wrote, no single “magic bullet.” Some of those strategies will be the same ones we’ve used for years: discovery and development of new drugs, careful monitoring for new resistant strains. Others, however, are new approaches, such as using drug cycling to guide bacterial evolution in the direction doctors want it to go (vide infra).
____
____
Surveillance of AMR:
Antimicrobial resistance (AMR) is a leading worldwide threat to the wellbeing of patients, and the safety and quality of health care. Although they have been available only for the past 80 years, antibiotics are accepted as an essential part of everyday health care, both in hospitals and in the community. Indeed, many current medical practices, such as major abdominal surgery, cancer chemotherapy, organ transplantation, joint replacement and neonatal care, are not possible without their use – without antimicrobials, mortality and morbidity during these procedures would be too great. AMR is developing at an alarming pace. Resistance often occurs within months of the release of new antimicrobials, and the resistance incidence rates outstrip drug discovery and the development of new antibiotics. The world is now facing the very real possibility of a return to non‑treatable infections, severe limitations on medical procedures and escalating healthcare costs. Surveillance and reporting of AMR and antibiotic usage is central to their prevention and containment. Data generated through surveillance of AMR and antibiotic usage are complementary and fundamental to everyday practices. At the local level, the data are used to formulate recommendations for rational antibiotic use and standard treatment guidelines. At a national level, data on resistance and antibiotic use inform policy decisions, such as antibiotic guideline development or revision, and identify priorities for public health action, such as education campaigns or regulatory measures. Without comprehensive and coordinated surveillance systems, efforts to prevent and contain AMR may be misdirected and inefficient, whereby poor practices such as inappropriate therapy result in wasted limited resources, and harm and human suffering through the inability to provide an effective drug to patients in need. Globally, there are a number of different programs for the surveillance of both AMR and antibiotic usage.
_
The need for surveillance of antibiotic resistance:
Successful treatment of serious infections requires timely administration of effective chemotherapeutic agents. While some infections (e.g. TB, whooping cough and gonorrhoea) are caused by a single pathogen, the majority of infections, such as those affecting the skin and soft tissues, the upper and lower respiratory tracts, the urinary tract, meningitis and sepsis are caused by a range of pathogens. Hence, clinical decisions about empirical treatment require knowledge of the likely pathogen(s) and the likely susceptibility of these pathogens to antibiotics. Such knowledge is gained in part by clinical experience over time, but more objectively and robustly through surveillance. The US Centers for Disease Control and Prevention (CDC) have defined surveillance as ‘The on-going systematic collection, analysis and interpretation of health data essential to the planning, implementation and evaluation of public health practice, closely integrated with the timely dissemination of these data to those who need to know’. Put more succinctly, surveillance is the generation and timely provision of information to inform decision-making and action. Implicit in these definitions is the fact that undertaking surveillance requires a readily available source of data. For surveillance of antibiotic resistance, the essential core data are generated by microbiology laboratories that routinely identify and determine the susceptibility or resistance of bacteria isolated from clinical specimens. These results are stored in the laboratory computer system and if accessed, collected and analysed, can inform as to the degree of antibiotic resistance seen in different bacterial species or isolates from different types of infection. Changes or variation in antibiotic resistance either geographically or over time can also be monitored.
_
Centralised surveys to validate routine data offer a practical approach:
It is easiest to count resistance rates of bacteria received at laboratories, but these organisms form a biased sample because (a) laboratory requesting varies greatly among clinicians; (b) some diseases (such as chronic obstructive airways disease) are more likely to generate laboratory specimens than others (such as pneumonia); (c) some age groups, particularly the elderly, are more likely to have specimens taken than others; and (d) primary care specimens are usually sent only from patients who have failed to respond to empirical treatment or who have comorbidities. Ideally resistance should have a clinical denominator (number of infected patients) not a laboratory one (number of isolates), but this is not easy except in uncommon diseases such as tuberculosis in the United Kingdom. If surveillance is based on isolates submitted to laboratories either routine susceptibility results can be collected or the isolates can be sent to a central laboratory for testing. Using routine results exploits data that exist already in sufficient quantities for relation to prescribing and population denominators. However, the quality of these data is patchy if, as in Britain, laboratories use different methods and do not routinely speciate many fermentative Gram negative bacilli. Few antibiotics are tested against all isolates, and “second line” antibiotics are tested only against those with an index resistance, giving a very biased sample. Finally, unless the data are analysed with respect to antibiogram phenotypes, anomalies and new mechanisms cannot reliably be recognised. Centralised testing, or testing to an agreed protocol by sentinel laboratories, allows standardised methods and measurement of levels of resistance. It also allows early detection of those resistances that accrue and can be linked to molecular studies to identify resistance mechanisms and monitor the spread of their encoding genes. However, centralised testing is limited by throughput and the sentinels may forma biased sample. Both routine data gathering and centralized surveys are undertaken. International cooperation on surveillance of resistance is desirable, not least to determine the extent to which different national prescribing practices translate into different resistance rates. To this end, the World Health Organisation is establishing networks of surveillance networks. At the other extreme, and critically, good local surveillance is needed to inform empirical treatment and to help individual hospitals manage their resistance problems.
_
Using whole-genome sequencing (WGS) to investigate transmission of resistant bacteria:
It is becoming increasingly clear that the discriminatory power of WGS to differentiate between epidemiologically related and unrelated isolates within clones is greatly enhancing our ability to investigate clusters and putative outbreaks of antibiotic-resistant infections. For example, using conventional outbreak investigation methods, a hospital infection control team noted a putative outbreak of MRSA on a special care baby unit (SCBU), where 12 infants colonized with MRSA over a six-month period were suspected but not categorically proved to be linked. WGS analysis of MRSA isolates from the SCBU and elsewhere not only confirmed that the cluster did indeed comprise an outbreak, but that it was more extensive than originally envisioned, with transmission of the outbreak strain between mothers on a postnatal ward and in the community also being found. Conversely, in another study, apparent instances of patient-to-patient transmission of MRSA in an adult intensive care unit identified by spa typing of isolates from patients with overlapping stays were found to have been incorrectly identified when subsequent WGS analysis showed that the isolates from ‘epidemiologically linked’ patients were in fact genetically distinct. Various studies are being published showing that WGS has the potential to unravel the complex epidemiology of antibiotic-resistant Gram-negative bacteria infections, including the emerging and critically important problem of infections caused by carbapenem-resistant organisms.
_
Importance of AMR surveillance:
Routine antimicrobial surveillance is an invaluable tool for nations, regions, and local facilities, because it is essential to ensure accurate information in order to establish and modify treatment guidelines and to aid in the prescription of appropriate empirical antimicrobial therapy. Although international and national studies inform and educate, they are of restricted benefit for decision making with regard to individual patients; such decisions instead must essentially rely on local intelligence from local surveillance. In contrast, routine surveillance of antimicrobial use reveals trends in dosing and allows comparisons of antimicrobial use data with antimicrobial resistance data that can provide important insights into the influence of particular use patterns on resistance. Unlike resistance-surveillance work, in which local data have little international significance, the results of antimicrobial use surveillance are frequently applicable internationally. The findings from both types of surveillance can point to the need for specific initiatives in prescribing practices, which may then themselves be evaluated longitudinally by use of the same methods. Given the relevance of antimicrobial resistance and antimicrobial use surveillance studies in the modern world of infection management, it is clear that they are here to stay. To deliver the best and most useful information into the future, surveillance studies need to maintain a rich mix of the large and small, the global and the local. Surveillance studies provide important information that allows for the identification of trends in pathogen incidence and antimicrobial resistance, including identification of emerging pathogens at national and global levels. Routine surveillance is critical for creating and refining approaches to controlling antimicrobial resistance and for guiding clinician decisions regarding appropriate treatment. The traditional approach has been to monitor pathogen antimicrobial susceptibility; numerous large studies have been performed, and their designs have evolved over time. Longitudinal studies are particularly useful because important information can be obtained by comparing data over time. Another approach to surveillance, that of monitoring antimicrobial use, can help to identify trends in dosing, to prevent the development of resistance. Several studies have incorporated this approach into their methods, and both large and small studies have attempted to correlate antimicrobial use data with antimicrobial resistance data. Overall, care must be taken to coordinate programs for optimal utilization of resources, to avoid duplication of effort. Information obtained from antimicrobial surveillance studies is important for establishing trends in pathogen antimicrobial resistance and for identifying emerging pathogens at the national and global levels. This information enables the development of targeted approaches to help control antimicrobial resistance. In addition, surveillance data regarding variations in trends among different countries, regions, and local facilities (e.g., hospitals and nursing homes) can help to guide treatment decisions so that clinicians may avoid initiating inappropriate antimicrobial therapy. Further, because large antimicrobial surveillance studies are often performed on a routine basis, comparisons of data obtained over time may aid in the assessment of the effectiveness of interventions.
_
Data from national and global surveillance studies indicate that the incidence of antimicrobial-resistant pathogens is increasing. For example, data obtained from 1997 to 2003 by the National Nosocomial Infections Surveillance (NNIS) System that illustrate the increasing incidence of Pseudomonas aeruginosa isolates, from patients in US intensive care units (ICUs), that are resistant to third-generation cephalosporins, quinolones, and imipenem. This increase in antimicrobial resistance, which compromises the use of previously effective treatments for antimicrobial infections, is of great concern for public health. Acquired carbapenemases are emerging resistance determinants in Gram-negative pathogens, including Enterobacteriaceae, Pseudomonas aeruginosa and other Gram-negative non-fermenters. A consistent number of acquired carbapenemases have been identified during the past few years, belonging to either molecular class B (metallo-β-lactamases) or molecular classes A and D (serine carbapenemases), and genes encoding these enzymes are associated with mobile genetic elements that allow their rapid dissemination in the clinical setting. Therefore, detection and surveillance of carbapenemase-producing organisms have become matters of major importance for the selection of appropriate therapeutic schemes and the implementation of infection control measures.
______
______
Prevention, containment and management AMR:
_
_
AMR management include:
_
WHO response:
Tackling antibiotic resistance is a high priority for WHO. A global action plan on antimicrobial resistance, including antibiotic resistance, was endorsed at the World Health Assembly in May 2015. The global action plan aims to ensure that the prevention and treatment of infectious diseases with safe and effective medicines continues.
The global action plan has 5 strategic objectives:
•To improve awareness and understanding of antimicrobial resistance
•To strengthen surveillance and research
•To reduce the incidence of infection
•To optimize the use of antimicrobial medicines
•To ensure sustainable investment in countering antimicrobial resistance.
_
To ensure you can respond effectively on every level, you need commitment from all those involved, complete and reliable information, and relevant, timely tools.
1. Prevention
•Infection control
-Screening and active surveillance
-Hygiene policy
•Antimicrobial Stewardship Programs
•Environmental control
2. Treatment
•Differentiation of bacterial from viral infection
•Informed decisions for relevant antibiotic use
-Rapid tests
-Reliable identification of bacteria
-Antimicrobial resistance testing
•Flexible testing methods for different healthcare/veterinary situations
-Easy to read and interpret results
3. Outbreak management & tracking
•Rapid detection of resistance mechanisms
•Complete epidemiological data tracking
•Good information flow
4. Education
•Collaborative action at the local, national and international levels
•Continuing education for healthcare professionals
•Accurate, easy-to-access patient education
_____
Prevention of AMR:
Ways to prevent transmission of all organisms, including antibiotic resistant bacteria, are:
•Wash hands before and after food handling, going to the toilet and changing nappies.
•Cover your nose and mouth when coughing and sneezing.
•Use tissues to blow or wipe your nose.
•Dispose of tissues properly, either in the rubbish or toilet.
•Do not spit.
•Stay at home if you are unwell and cannot manage your normal requirements of the day.
•Do not send children to child care, crèche or school if they are unwell.
•If you are prescribed antibiotics, take the entire course – do not stop because you are feeling better.
•If you continue to feel unwell, go back to the doctor.
•Avoid use of products that advertise they contain antibiotics, or are antibacterial or antimicrobial, unless advised to do so by your health professional.
_
Antibiotic resistance is accelerated by the misuse and overuse of antibiotics, as well as poor infection prevention and control. Steps can be taken at all levels of society to reduce the impact and limit the spread of resistance.
1. The general public can help by:
•Preventing infections by regularly washing hands, practicing good food hygiene, avoiding close contact with sick people and keeping vaccinations up to date
•Only using antibiotics when prescribed by a certified health professional
•Always taking the full prescription
•Never using left-over antibiotics
•Never sharing antibiotics with others.
2. Health workers and pharmacists can help by:
•Preventing infections by ensuring hands, instruments and environment are clean
•Keeping patients’ vaccinations up to date
•When a bacterial infection is suspected, perform bacterial cultures and testing to confirm
•Only prescribing and dispensing antibiotics when they are truly needed
•Prescribing and dispensing the right antibiotic at the right dose for the right duration.
3. Policymakers can help by:
•Having a robust national action plan to tackle antibiotic resistance
•Improving surveillance of antibiotic-resistant infections
•Strengthening infection prevention and control measures
•Regulating and promoting the appropriate use of quality medicines
•Making information on the impact of antibiotic resistance available
•Rewarding the development of new treatment options, vaccines and diagnostics.
4. The agricultural sector can help by:
•Ensure that antibiotics given to animals – including food-producing and companion animals – are only used to treat infectious diseases and under veterinary supervision.
•Vaccinate animals to reduce the need for antibiotics and develop alternatives to the use of antibiotics in plants.
•Promote and apply good practices at all steps of production and processing of foods from animal and plant sources.
•Adopt sustainable systems with improved hygiene, biosecurity and stress-free handling of animals.
•Implement international standards for the responsible use of antibiotics, set out by OIE, FAO and WHO.
5. The healthcare industry can help by:
•Investing in new antibiotics, vaccines, and diagnostics.
_____
Alliance for the Prudent Use of Antibiotics (APUA):
The Alliance for the Prudent Use of Antibiotics (APUA) is a non-profit organization founded in 1981 by Stuart B. Levy, Professor of Medicine at Tufts University and headquartered in Boston, Massachusetts. APUA’s mission is to strengthen society’s defenses against infectious disease by promoting appropriate access and use to antimicrobial agents (antibiotics, antivirals, antimalarials etc.) and controlling antimicrobial resistance on a worldwide basis. APUA has a network of affiliated chapters in over 50 countries, and conducts applied antimicrobial resistance research, education, capacity building and advocacy at the global and grassroots levels. Wide-scale misuse of antibiotics and other antimicrobials and related resistance to these drugs is challenging infectious disease treatment and health care budgets worldwide. Antimicrobials are uniquely societal drugs because each individual patient use can propagate resistant organisms. APUA’s provides information to individuals, doctors and policy makers aimed at preserving the power of these agents by preventing infection, reducing drug resistance and increasing the effectiveness of treatment for infectious diseases, including acute bacterial diseases, tuberculosis, AIDS and malaria.
_______
Interventions against AMR:
Interventions against AMR bacteria may aim to reduce the transmission of existing resistant strains, or prevent the development of further resistance. Arguably, hand hygiene has been the primary strategy employed aiming to reduce transmission, and antibiotic stewardship the cornerstone for the slowing or prevention of resistance development. While many AMR infection prevention and control strategies exist (primarily for the hospital setting), evidence of their effectiveness from well conducted trials is lacking. There remains uncertainty over the efficacy of infection control strategies for a number of reasons. Results from trials may be contradictory, and often evaluate different and therefore incomparable, intervention strategies. The effectiveness of strategies may differ between settings, for example by prevalence or specialty, and it is not obvious how findings should be generalized. In addition, often many infection prevention and control interventions are employed at the same time, making it difficult to determine which components are having an effect. It is extremely rare that interventions are rigorously assessed in clinical trials, and those that have been are for general infection prevention measures not specifically targeted against AMR pathogens. However, while there are few clinical trials of specific interventions against AMR, the overall infection prevention and control literature is vast. Selected examples from the published literature, many of which are systematic reviews across the broad range of interventions, are provided here. Antimicrobial stewardship programmes are increasingly being advocated as a means of improving the quality of prescribing. Recent Cochrane reviews evaluating stewardship interventions include: that by Davey et al assessing the evidence for interventions aiming to improve antibiotic prescribing practices for hospital inpatients; the assessment of evidence on prophylactic use of antibiotics to reduce morbidity and mortality in ventilated newborn infants by Inglis et al; and the assessment of antibiotic prophylaxis to reduce respiratory tract infections and mortality in adults in intensive care by Liberati et al. In order to address the need for increased understanding of both antibiotic usage and resistance patterns, the English Surveillance Programme for Antimicrobial Utilization and Resistance (ESPAUR) was established in 2013. The Programme is bringing together antimicrobial surveillance in both primary and secondary care settings, developing quality measures and methods to monitor unintended outcomes of antimicrobial stewardship and behaviour-based interventions. Hand hygiene as an intervention has been subject to many studies and therefore has a large literature. Stone et al attempted to assess the effect of increased hand hygiene procurement used in NHS trusts in England and Wales, on the observed reduction in MRSA bacteraemia and C. difficile infection (CDI), accounting for other interventions. Associations between increased alcohol hand rub and reduced MRSA bacteraemia, and increased liquid soap use and reduced CDI were found. However, there were a number of limitations in this ecological study, particularly the inability to obtain data on mupirocin usage for decolonization of MRSA and antimicrobial prescriptions data, which is clearly associated with CDI. In an assessment of a behavioural intervention to improve hand hygiene compliance, Stone et al found peer group audit and feedback significantly improved compliance, however effects waned towards pre-study levels over time. Both national and international guidance identifies hand hygiene as a key component in the reduction of AMR.
_
While new antibiotic options are critically needed, other approaches also have an important role. Both ensuring judicious use and shortening the duration of treatment can reduce antimicrobial use and reduce the development of AMR. Similarly, proven public health interventions such as access to clean water and sanitation and hospital infection control can prevent bacterial infections and obviate the need for some antibiotic use.
_
Public misunderstanding:
The public tends to think that it is the person taking the drugs who becomes resistant to their effects, not the microbes. A research published by the World Health Organisation showed that three-quarters of people in poor and middle-income countries misunderstood the problem that way. A survey carried out earlier in 2015 by the Wellcome Trust suggested a similar prevalence of misunderstanding in Britain. Such ignorance has consequences. If you know that resistance is an attribute of the bacteria, then using drugs rarely but definitively makes sense. Do not use them when not needed; when you do use them, use them in such a way as to kill off all the bacteria, rather than leaving behind a small resistant rump. If you mistakenly think resistance is an attribute of people, on the other hand, you will have no compunction about using antibiotics, provided they seem to have some effect. And you will not think twice about stopping the course when the symptoms subside, rather than carrying it on until all the bacteria are gone. These problems are at their worst in places where antibiotics are easily bought over the counter. It is reasonable to assume that public-awareness campaigns might put things right. But this seems a little optimistic. Even when prescriptions are needed and experts are involved things still go wrong. In America some 40 million people are prescribed antibiotics for respiratory problems every year. In 2013 a paper published in the Journal of Antimicrobial Chemotherapy estimated that two-thirds of those people may well not have needed the antibiotics they got. Some of this is down to “pester power”: having gone to the doctor, a patient wants something tangible to show for it, even if his sore throat is probably viral and antibiotics will do him no good. Sometimes, though, it is the other way around. If a doctor cannot be sure of the cause, prescribing an antibiotic may help. The real chance of healing a specific person outweighs the imperceptible increase to the threat of bacterial resistance.
________
Harnessing the Immune System:
Vaccines:
Microorganisms do not develop resistance to vaccines because a vaccine enhances the body’s immune system, whereas an antibiotic operates separately from the body’s normal defenses. Furthermore, if the use of vaccines increase, there is evidence that antibiotic resistant strains of pathogens will decrease; the need for antibiotics will naturally decrease as vaccines prevent infection before it occurs. Laxminarayan and colleagues estimated that improved vaccination coverage for Streptococcus pneumoniae could avert 11.4 million antibiotic days per year in children under 5 years of age worldwide. However, new strains that escape immunity induced by vaccines may evolve; for example, an updated influenza vaccine is needed each year. Vaccines and antibiotics have significantly contributed to improve health and also to increase the longevity of human beings. The fast-acting effect of antibiotics makes them indispensable to treat infected patients. Likewise, when the causative agent of the infection is unknown and in cases of super-infections with different species of bacteria, antibiotics appear to be the only therapeutic option. On the contrary, vaccines are usually not efficacious in people already infected and their action is generally limited to a much narrowed range of pathogens. However, vaccines have contributed to the eradication of some of the most deadly infectious agents worldwide, can generate immunity to infections lasting for several years or life-long, and are able to induce herd immunity. Nonetheless, infectious diseases are still among the leading causes of morbidity and mortality worldwide. This is mainly owing to the emergence of bacterial resistance to antibiotics and the lack of efficacious medications to treat several other infectious diseases. Development of new vaccines appears to be a promising solution to these issues. Indeed, with the advent of new discovery approaches and adjuvants, today is possible to make vaccines virtually against every pathogen. In addition, while vaccine-resistant bacteria have never been reported, accumulating literature is providing evidence that vaccination can reduce the raise of antibiotic resistant strains. Vaccines capable of preventing bacterial infection, disease, or both circumvent the problem of AMR and have been successfully developed for several bacterial pathogens. However, the development of vaccines for health care–associated bacterial pathogens has been challenging, due to inadequate understanding of immune correlates of protection, complex pathogenic mechanisms, and extensive strain and antigenic variability. In this regard, several staphylococcal vaccine development programs have failed at the phase 3 stage of clinical trials, despite promising preclinical and early clinical data. However, a number of vaccine candidates for health care–associated infections remain in clinical development and, if successful, would likely be deployed in targeted populations of at-risk individuals.
_
Passive infusion of monoclonal antibodies provides additional options for treatment and prevention. Monoclonal antibodies are being developed for use in combination with antibiotics in severely ill patients with certain bacterial infections (e.g., S aureus and P aeruginosa), as well as for prophylactic use. The low toxicity and long serum half-lives of certain monoclonal antibodies as well as the absence of conventional, drug-mediated selective pressure when they are used as antimicrobials make them attractive options, especially for prophylaxis. Monoclonal antibodies are particularly promising for patient populations with suboptimal responses to vaccination because of immune compromise, immune senescence, or other conditions. In addition, bispecific antibodies that can simultaneously bind pathogens and activate T cells have been developed for use against tumors and virus-infected cells. In the future, such innovations could be adapted to target bacterial infections. Furthermore, considerable progress has been made in identifying the signalling pathways and receptors of the innate immune system. Investigators have identified new potentiators of innate immunity that may be effective as vaccine adjuvants or directly as therapeutic modalities. Such immune enhancements could lessen the required antibiotic dose, treatment duration, or both, thereby decreasing selective pressure toward resistance. Some prospects include broadly active innate immune–based strategies such as defensins, bactericidal/permeability–inducing protein, engineered γ-core motif peptides or peptidomimetics, complement, components of mucosal secretions including surfactant, and inflammation resolving mediators.
______
Antimicrobial/ antibiotic stewardship:
The appropriate use of antibiotics — often called antibiotic stewardship — can help preserve the effectiveness of current antibiotics, extend their life span and protect the public from antibiotic-resistant infections. Many hospitals and medical associations have implemented new diagnostic and treatment guidelines to ensure effective treatments for bacterial infections and reduce inappropriate use of antibiotics. Antimicrobial stewardship program is a series of policies introduced by the government for improving antimicrobial prescribing behaviors of doctors. It mainly includes leadership commitment and accountability, and key supporting groups implement policies for optimal antimicrobial use and interventions to avoid antimicrobial resistance. The policies support the implementation of facility-specific treatment recommendations. The antimicrobial stewardship programs were recommended by the Centers for Disease Control and Prevention (CDC) for improving the antimicrobial prescription in hospitals (Centers for Disease Control and Prevention 2013; Centers for Disease Control and Prevention 2014). The World Health Organization (WHO) has recommended training for medical undergraduates regarding the prudent prescription of antimicrobials (World Health Organization 2012). It is necessary that our future doctors are better equipped with better knowledge of antimicrobial use and resistance. Unlike the senior doctors or infectious specialists who have a large amount of experience in anti-infection treatment, the junior doctors usually have limited knowledge and skills to reduce the potential risk of antimicrobial resistance (Charani et al. 2010). Therefore, antimicrobial stewardship efforts should be made to standardize the prescribing behaviors of our future doctors (Hecker et al. 2003; Owens et al. 2004; Paterson 2006). However, thus far, less attention has been given to future doctors during their medical colleges. Studies of medical students from various countries have documented students’ perceptions on antimicrobial stewardship and students’ feelings about their education regarding antimicrobial use and resistance (Abbo et al. 2013; Afzal Khan et al. 2013; Dyar et al. 2013; Ibia et al. 2005; Luther et al. 2013; Minen et al. 2010; Pulcini et al. 2015; Thriemer et al. 2013). Previous studies on antimicrobial education in the United States reported that there is an obvious gap among the medical students from different US medical schools in terms of choosing study references, preparedness for prescribing antimicrobials, and perceptions of knowledge regarding antimicrobials (Abbo et al. 2013; Ibia et al. 2005; Minen et al. 2010). Incorporating principles of antimicrobial stewardship and appropriate use into undergraduate and postgraduate medical education can be implemented and is under consideration by the Government of India. A more difficult problem is that of regulating the sales of substandard and illegitimate antimicrobials, the extent of which is poorly quantified.
_
Antibiotic stewardship has many elements:
1. Use a procalcitonin level as a biomarker for infection to avoid unnecessary antibiotic use, as has been shown to be successful in nearly every well-controlled trial
2. Short courses of antibiotics are virtually always effective in well-controlled trials.
3. Switch antibiotics from intravenous (IV) to oral formulations to hasten discharge and reduce risks associated with IV catheters. This switch is easily done with many antibiotics (linezolid, metronidazole, fluoroquinolones, some cephalosporins, fluconazole, etc.).
4. Use colistin carefully. Colistin, available since 1961, is increasingly needed but is saddled with dosing errors because the recommendations in the package insert are wrong.
5. Avoid antibiotic redundancy, as illustrated by the report that 23% of 782,821 patients were given metronidazole on top of another agent for anaerobic bacteria.
_
Antibiotic combinations:
Rationale:
A patient takes a single antibiotic, and all bacteria resistant to that drug survive. A patient takes several antibiotics, and few bacteria survive because most are not resistant to all of the drugs. This is especially applicable for infections caused by multiple pathogens affecting the skin and soft tissues, the upper and lower respiratory tracts, the urinary tract, meningitis and sepsis. Multiplicity of pathogens needs combination of antibiotics although tuberculosis caused by single pathogen also warrant combination of antibiotics.
_
Is administration of combination therapy needed to prevent resistance?
To increase the likelihood of adequate coverage, the empirical antimicrobial regimen for VAP or other ICU-acquired infections in patients at risk for multidrug-resistant GNB usually combines a broad-spectrum beta-lactam with anti-pseudomonal activity and either an aminoglycoside or an anti-pseudomonal fluoroquinolone. However, when both agents are active, the benefit of combination therapy over adequate monotherapy has not been proven in terms of clinical cure or microbiological eradication. Convincing evidence is similarly lacking to support the routine use of antimicrobial combinations (including a beta-lactam) as definite regimen in an attempt to prevent the emergence of resistance under therapy. In P. aeruginosa infections, adding an aminoglycoside to an effective beta-lactam does not prevent from the emergence of beta-lactam resistance, including in patients treated with imipenem. In Enterobacteriaceae, the main mechanism of acquired beta-lactam resistance under therapy is chromosomal AmpC derepression. In a prospective cohort of 218 patients infected with natural AmpC producers and receiving 3GC (third generation cephalosporin), the emergence of 3GC resistance was observed in 11 cases (5%): combining 3GC with an aminoglycoside or a fluoroquinolone did not significantly reduce the rate of mutant selection. Therefore, once susceptibility testing results are known, monotherapy with the most active beta-lactam could be considered, with high-dosing regimen and optimized administration. Clinical data remain scarce for infections due to multidrug-resistant A. baumannii, although in vitro studies indicate that combining colistin with rifampicine, carbapenems or tigecycline may be effective to prevent the emergence of colistin-resistant mutants.
_
High dose short duration:
Dr Arnold Louie from the University of Florida presented a roadmap of PK/PD principles for decreasing the development of drug resistance in antimicrobials. He suggested combating resistance for single drug regimens by hitting the bacteria hard and fast. Louie recommends using higher dosages for a shorter duration to rapidly reduce the total population and cut the use of antibiotics.
_
Rotating Use of Antibiotics:
The most recent intervention in antibiotic prescribing has been renewed interest in rotating use, or cycling, of antibiotics. Over 20 years ago, in a series of studies at the Minneapolis Veterans’ Administration Hospital, the substitution of amikacin for gentamicin and tobramycin as the aminoglycoside of choice produced sustained decreases in the prevalence of aminoglycoside-resistant gram-negative bacilli. The higher serum levels of amikacin, and the infrequent appearance in U.S. hospitals of amikacin-modifying enzymes that could confer amikacin resistance in gram-negative bacilli, were the underpinnings of the success of this strategy. The more recent reports on cycling describe replacement (or switch) therapy for empiric antibiotic choices. Replacing ceftazidime with ciprofloxacin for empiric treatment of suspected gram-negative bacterial infections in a cardiac surgery ICU was associated with decreased incidence of ventilator-associated pneumonia and bacteremia caused by antibiotic-resistant gram-negative bacilli. In another hospital, use of beta-lactam/beta-lactamase inhibitor combinations to replace use of third-generation cephalosporins and clindamycin was associated with decreased rates of colonization by VRE; a follow-up study reported that these formulary manipulations were associated with decreasing numbers of patients from whom methicillin-resistant S. aureus and ceftazidime-resistant K. pneumoniae were cultured but increased rates of resistant Acinetobacter. Rotating use of fourth-generation cephalosporins, quinolones, carbapenems, and beta-lactam/beta-lactamase inhibitor combinations is being studied in several hospital ICUs. Cycling of antibiotics is most likely to be effective for limited periods in closed environments, such as ICUs, but this approach requires careful microbiologic monitoring because of the monotonic selective pressure of a single agent and the possible emergence of resistance to unrelated classes of drugs caused by genetic linkage of resistance mechanisms. As the size of the patient population under study increases, availability of various classes of drugs may be more effective at reducing the risk of emergence of resistance and may be a better strategy than cycling.
_
Alternating therapy (vide infra):
Alternating therapy is a proposed method in which two or three antibiotics are taken in a rotation versus taking just one antibiotic such that bacteria resistant to one antibiotic are killed when the next antibiotic is taken. Studies have found that this method reduces the rate at which antibiotic resistant bacteria emerge in vitro relative to a single drug for the entire duration.
_
WHO guidance says no routine post-surgery antibiotics:
New guidelines on surgical site infections (SSIs) from the World Health Organization (WHO) say antibiotics should be used to prevent infection before and during surgery, but not after. While preventing infections from happening in the first place is important for reducing the spread of drug-resistant superbugs, the recommendation that antibiotics be used to prevent infections only before and during surgery, and not after, is seen as a crucial measure in efforts to reduce the overuse of antibiotics. Overuse, like post-operatively, increases the selective pressure for the emergence of drug-resistant infections.
_
Syndromic Approach for Empirical Therapy of common infections:
Empirical or presumptive anti-infective therapy is based on a clinical diagnosis combined with evidence from the literature and from the educated experience of the probable pathogens causing the infection. To optimize an accurate microbiological diagnosis, clinicians should ensure that diagnostic specimens are properly obtained and promptly submitted to the microbiology laboratory, preferably before the institution of antimicrobial therapy. All attempts should be made to establish diagnosis of the patients based on the facilities available to the treating doctor and affordability of the patients. Definitive therapy depends on the microbiologic diagnosis by isolation or other direct evidence of pathogen. According to WHO, presumptive treatment is a one-time treatment given for a presumed infection in a person, or group of people, at high risk of infection. Presumptive treatment is prescribed typically while waiting for the culture report or in situations where the facilities for doing these tests is not available, is difficult or not cost effective or is impractical. However in certain situations the empirical therapy prescribed as prophylaxis also (e.g. surgical prophylaxis, high prevalence, repeated risk of exposure). The syndromic approach is based on the presence of consistent groups of symptoms and easily recognized signs caused by a single pathogen or a mixture of pathogens.
Before starting presumptive therapy ensure the following:
1. Send and follow up on standard investigations for all suspected infections for correct and accurate diagnosis and prognosis.
2. Antibiotics should be started only after sending appropriate cultures if facilities are available. Similarly any change in antibiotic must be guided by sensitivity profile.
3. Assess the factors affecting activity of antimicrobials such as renal excretion, interactions and allergy before prescribing antibiotics.
4. Review of antibiotic therapy must be done daily and the therapy escalated or deescalated accordingly especially after the culture reports are available.
Empirical Therapy is justified in patients with life threatening infections, in ICU settings and while awaiting results of culture. The timing of initial therapy should be guided by the patient’s condition and urgency of the situation. In critically ill patients e.g. patients in septic shock or bacterial meningitis therapy should be initiated immediately after or concurrently with collection of diagnostic specimens. In other conditions where patient is stable, antimicrobial therapy should be deliberately withheld until appropriate specimens have been collected and submitted to the microbiology laboratory e.g. when treating a patient of osteomyelitis or sub-acute endocarditis. Premature usage of antimicrobial in such cases can preclude opportunity to establish a microbiological diagnosis, which is critical in the management of these patients.
__
Practice points for prescribers:
•Consider the nature and severity of the infection and the person’s immune status, and prescribe antibiotics when benefits are likely to be substantial.
•Refer to the antibiotic guidelines for the recommended duration of antibiotic therapy, or, where applicable, local evidence-based guidelines.
•When prescribing antibiotics, specify duration of treatment.
•Bear in mind that the shorter the duration of treatment, the lower the selection pressure for resistance is in the patient.
•Do not provide repeat prescriptions unless required (consider removing the default for repeat prescription in medical software).
•Consider including an expiry date on prescriptions to prevent them from being filled after anticipated resolution of infection.
• Advise patients to take the antibiotics as directed, even if this means there are antibiotics remaining.
•Evaluate the need to continue, revise or stop antibiotic treatment based on clinical response and available microbiological data.
_
The public also plays a role in antibiotic stewardship. You can help reduce the development of antibiotic resistance by taking the following steps:
•Use antibiotics only as prescribed by your doctor.
•Take the appropriate daily dosage and complete the entire course of treatment.
•If you have an antibiotic prescription, ask your doctor what you should do if you forget to take a dose.
•If for some reason you have leftover antibiotics, throw them away. Never take leftover antibiotics for a later illness. They may not be the correct antibiotic and would not be a full course of treatment.
•Never take antibiotics prescribed for another person.
•Don’t pressure your doctor to give you an antibiotic prescription. Ask your doctor for advice on how to treat symptoms.
•Practice good hygiene. Wash your hands regularly with soap and water, especially after using the toilet, before eating, before preparing food and after handling fresh meat. Wash fruits and vegetables thoroughly, and keep kitchen work surfaces clean.
•Make sure you or your children receive recommended vaccinations. Some recommended vaccines protect against bacterial infections, such as diphtheria and whooping cough (pertussis).
•If you think you may have penicillin allergy, talk to your doctor about getting an allergy skin test. Research has shown that penicillin or other antibiotic allergies may be over reported. Ruling out an antibiotic allergy can help your doctor prescribe the most appropriate antibiotic when it’s needed.
_
Social media proves effective as a tool for antimicrobial stewardship: a 2016 study:
A new study from the University of Chicago Medicine examines the use of social media platforms to inform young physicians about proper use of antimicrobial agents such as antibiotics. A study by Jennifer Pisano, MD, and colleagues appearing in the American Journal of Infection Control, finds that social media platforms – including Facebook and Twitter — provide an effective method to reinforce antimicrobial stewardship programs (ASP) and encourage the use of ASP resources to promote antimicrobial mindfulness among internal medicine residents. The strategy pioneered by the researchers successfully directed medical residents to the appropriate use of clinical pathways. Over the course of six months, 55 medical residents received Facebook posts and tweets of basic information promoting both educational tools and clinical pathways located on the researchers’ hospital’s ASP website. The medical residents also received identical infectious disease and antibiotic knowledge “trivia questions,” as well as interspersed questions Participants’ knowledge of how to use the ASP website increased from 70 percent to 94 percent, while these residents’ antibiotic knowledge also improved. Crucially, use of relevant clinical pathways sometimes, frequently, or always increased from 33 percent to 61 percent (P = 0.004).
______
ASP in rehab facility reduces antibiotic use and resistance: a 2016 study:
The results of a small, single-center study suggest that implementing an antibiotic stewardship program (ASP) based on infectious disease (ID) consultation in a rehabilitation facility can reduce antibiotic use and antimicrobial resistance without affecting patient outcomes. The quasi-experimental study, reported in Infection Control and Hospital Epidemiology, compared the periods before and after an ASP was implemented at a 150-bed rehabilitation facility in northern Italy specializing in spinal cord injuries (SCI). The authors note that patients cared for in SCI rehab hospitals are prone to infections dues to several factors—including bladder catheterization, invasive procedures, and pressure sores—and that antibiotic over-prescribing in these facilities is a recognized problem. During the study period (January 2011 to December 2014), overall antibiotic consumption at the facility decreased by 48%, dropping from 42 defined daily doses (DDD) per 100 patient days in 2011 to 22 DDD in 2014. Specifically, the use of carbapenems dropped by 97% and fluoroquinolone use dropped by 92%. The use of aminoglycosides, tetracycline, clindamycin, and macrolides also declined. The consumption of third-generation cephalosporins remained stable. At the same time, the researchers found that the incidence of Clostridium difficile fell from 3.6 cases per 10,000 patient days in 2011 to 1.2 cases in 2014. In addition, they observed a significant decrease in drug-resistant bacteria. The prevalence of extensively drug-resistant (XDR) strains declined from 55% to 12% in Pseudomonas aeruginosa and from 96% to 73% in Acinetobacter baumanni; the prevalence of extended-spectrum beta-lactamase (ESBL) producing strains dropped from 42% to 17% in Escherichia coli and from 62% to 15% in Proteus mirabilis; carbapenem-resistant strains fell from 42% to 17% in Klebsiella pneumoniae; and methicillin-resistant strains of Staphylococcus aureus decreased from 77% to 40%. The authors note that while their study is limited by the single-center design and long-term monitoring is needed, the results suggest that an ASP based on systematic ID consultation can be effective outside of acute-care hospitals.
_____
______
Management of AMR in hospital:
Control of antibiotic resistance requires aggressive implementation of several strategies: ongoing surveillance of resistance; molecular typing of isolates, usually using pulsed-field gel electrophoresis when rates of resistance increase; using hygiene controls to limit spread of single (clonal) strains and antibiotic controls to limit spread of multiple (polyclonal) strains of resistant bacteria; and enlisting administrative support. Monitoring adherence of health-care workers to control measures and feedback of individual and ward rates of hygiene adherence and antibiotic resistance are central components of health-care worker education and motivation. Current infection control strategies are aimed at the hygiene and antimicrobial engines that drive resistance. To fulfil the infection control prime directive, we must harness technology to improve and direct adherence to these strategies. Future approaches may control or eliminate the bacterial events that underlie evolution of resistance.
_
Hospital Antibiotic Policy must be implemented:
1. To curb the common misuse and overuse of antibiotics
2. Restricts the occurrence of antibacterial resistance among the hospital strains
3. Controls the spread of such infections to susceptible and critically ill patients in the hospital and the subsequent infection into the community.
4. Hospital Antibiogram should have a periodic summary of antimicrobial susceptibilities of local bacterial isolates submitted to the hospitals clinical microbiology laboratory. It is used by clinicians to assess local susceptibility rates, as an aid in selecting empiric antibiotic therapy, and in monitoring resistance trends over time within an institution.
_
Steps to Combat Antimicrobial Resistance in Outpatient Settings:
| Steps | Suggestions for implementation |
| Improve antibiotic prescribing | |
| Use current clinical guidelines to support rational and appropriate antibiotic prescribing |
Share unremarkable finding during the examination (e.g., “no inflammation” or “normal breathing”), while acknowledging the patient is sick. Determine the likelihood of a bacterial infection, especially for upper respiratory tract infections. Provide a specific diagnosis (e.g., “viral bronchitis” vs. “virus”). Weigh benefits vs. harms of antibiotics |
| Communicate with patients about when and why antibiotics may not be necessary | Explain that unnecessary antibiotic use can be harmful (e.g., adverse effects associated with antibiotic use, potential resistance development). Explain that treating viral infections with antibiotics does not work. Explicitly plan treatment of symptoms by describing the expected normal course of the illness, and instruct patients to call or come back if symptoms persist or worsen; consider providing care packages with non-antibiotic therapies |
| Educate patients if an antibiotic is needed | Encourage adherence. Discuss potential adverse effects |
| Create an office environment that promotes a reduction in antibiotic use | Start the process in the waiting room with videos, posters, and other materials. Hang posters in examination rooms to display a commitment to not prescribe antibiotics for viral infections Involve office personnel in the reinforcement of the physician’s messages |
| Prevent infections and the spread of resistant bacteria | |
| Ensure that all patients get recommended vaccinations. Prevent cross-transmission |
Provide pneumococcal and influenza vaccines (to help avoid secondary bacterial infections) which are particularly important. Counsel patients on how to avoid spreading or becoming infected with resistant pathogens in the community (e.g., methicillin-resistant Staphylococcus aureus). Follow recommendations for infections control in outpatient department. |
| Monitor antibiotic-resistant infections | |
| Report notifiable diseases | Report to the health department any diseases caused by bacteria on the list of urgent and serious pathogens; antibiotic-resistant strains of some bacteria (e.g., methicillin-resistant Staphylococcus aureus) are reportable in some states. |
| Be alert for treatment failures | Consider the possibility of antibiotic resistance in cases of treatment failure; obtain laboratory confirmation and notify local public health authorities in case of unusual or unexpected treatment failure. |
_
Reduce Inappropriate Antibiotic Use in Outpatients:
The abuse of antibiotics is well known and in large part reflects consumer demand because the patient expects to walk out of the clinic with a prescription for that viral respiratory tract infection. A Cochrane review of all methods to reduce antibiotic abuse in the clinic concluded that the “3-day prescription” was the only method with documented success. This means telling the patients with “sinusitis” that they probably have a viral infection that is likely to get better within 3 days, and providing a prescription that is dated 3 days later for use if the patient is not better or is getting worse at that time.
_
Many hospitals, nursing homes, and other healthcare facilities take special safety measures to help prevent AMR including:
Hand washing: This is the single most important way to prevent the spread of AMR. Healthcare workers are taught to wash their hands with soap and warm water or use an alcohol-based hand sanitizer before and after treating each patient. They are also taught to clean their hands after touching any surface and after removing protective clothing.
Protective clothing: Healthcare workers and visitors wear gloves, a gown if soiling of clothes is likely, and sometimes a mask when entering the room of a patient with a MDR infection. The gown is removed, and hands are washed with soap and warm water before leaving the room.
Careful use of Antibiotics: Using antibiotics only when needed, and for the shortest time possible, helps prevent the growth of more MDR bacteria.
Private Rooms: Patients with a MDR infection are placed in a private room or share a room with others who have the same infection.
Daily cleaning: All patient care items, equipment, and room surfaces are properly cleaned and disinfected every day. Check lists may be used by staff to ensure all areas were cleaned.
Vaccinations: Streptococcus pneumoniae, a type of bacterium that can cause pneumonia, often can be resistant to one or more antibiotics. Vaccination against Streptococcus pneumoniae can prevent these antibiotic resistant infections.
Monitoring: Hospitals and other healthcare facilities monitor the spread of MDR bacteria and educate caregivers on the best ways to prevent it.
_
Antimicrobial copper touch surfaces may help fight global threat of antibiotic resistance: a 2016 study:
It is accepted that hand hygiene, and surface cleaning and disinfection, are standard measures to prevent and control infections in hospitals, but more needs to be done to prevent the spread of pathogens by staff, visitors and patients touching contaminated surfaces. What is not always appreciated is that bacteria deposited and surviving on a surface can exchange genes—including those for antibiotic resistance—which can result in new, resistant strains. Professor Bill Keevil, Chair in Environmental Healthcare at the University of Southampton, is a leading expert on the hygienic properties of copper, and believes replacing frequently-touched surfaces with antimicrobial copper equivalents—teamed with good hygiene practices—could help address both the environmental spread of contamination and the rise of antibiotic resistance. Copper is a powerful antimicrobial with rapid, broad-spectrum efficacy against bacteria and viruses, and has been shown to kill disease-causing pathogens, including influenza A, E.coli and norovirus, and resistant bacteria including Methicillin-resistant Staphylococcus aureus (MRSA), Carbapenem-resistant Enterobacteriaceae (CRE) and Vancomycin-resistant enterococcus (VRE). It shares this benefit with a range of copper alloys—such as brasses and bronzes—forming a family of materials collectively called ‘antimicrobial copper’. ‘We’ve shown that antimicrobial copper touch surfaces produce a rapid kill of bacteria, viruses and fungi, usually within minutes,’ says Professor Keevil. ‘EPIC 3—the national, evidence-based guidelines for preventing HCAIs in NHS Hospitals in England—recognize high-touch surfaces made from antimicrobial copper harbor 80–90% fewer bacteria than equivalent, non-copper surfaces. A multi-center trial in the US further showed a concurrent 58% reduction in HCAIs in ICU rooms equipped with antimicrobial copper touch surfaces.’ Professor Keevil also observes that gene transfer between bacteria does not occur on copper because bacteria are destroyed rapidly and completely. This means the genes for antibiotic resistance can’t be exchanged, contributing to a reduced likelihood of new resistant strains emerging.
______
Beyond antibiotics – alternative strategies for prevention and treatment:
Understanding the scientific basis of antimicrobial resistance is essential to combating this public health threat. There is no single solution and several, synergistic, overlapping, and complementing approaches will be needed, with a strong overarching shared goal to ensure and sustain access to effective antimicrobial therapies. These include:
1. Drug Derivatives: Modification of known antimicrobial agents – new β-lactamases or efflux pump inhibitors.
2. Novel agents: Whole cell or target based antibiotic discovery using new tools such as combinatorial chemistry and genomics.
3. Anti-virulence Drugs: Antibodies or small molecules blocking or inhibiting virulence factors.
4. Bacteriophages or enzybiotics: delivery of bacteriophages or phage-lytic enzymes.
5. Evolutionary biology approaches: Aimed at targeting the ecology/evolution of AMR, including inhibitors of plasmid transfer of resistance, gene silencing antisense oligomers.
______
Use of Molecular mechanism of AMR to overcome resistance:
Knowledge of the molecular mechanisms of antibiotic resistance is essential for developing new approaches to overcome this problem. One possible approach is the development of inhibitors of resistance enzymes. These inhibitors can be administered as co-drugs with the antibiotics, thereby blocking resistance and rescuing the antimicrobial activity of the drugs. Another strategy to overcome resistance is to improve the delivery or otherwise enhance the accessibility of antibiotics to their sites of action. For example, liposomal preparations of hydrophobic antibiotics, such as ethambutol for treatment of mycobacterial infections, have been reported. Strategies could be developed to target virulence factors of pathogens instead of whole bacteria (e.g. develop drugs that target the plasmids containing resistance genes or drugs that target the adhesion of virulent bacteria to a tissue). All the alternative strategies to overcome resistance require expanded knowledge of the molecular mechanisms of antibiotic resistance, their origins and evolution, and their distribution throughout bacterial populations and genomes.
____
β-Lactamase inhibitor:
Beta lactamases are a family of enzymes involved in bacterial resistance to beta-lactam antibiotics. They act by breaking the beta-lactam ring that allows penicillin-like antibiotics to work. Strategies for combatting this form of resistance have included the development of new beta lactam antibiotics that are more resistant to cleavage, and the development of beta lactamase inhibitors. Although β-lactamase inhibitors have little antibiotic activity of their own, they prevent bacterial degradation of beta lactam antibiotics and thus extend the range of bacteria the drugs are effective against. The most important use of beta lactamase inhibitors is in the treatment of infections known or believed to be caused by gram-negative bacteria, as beta lactamase production is an important contributor to beta lactam resistance in these pathogens. In contrast, most beta lactam resistance in gram-positive bacteria is due variations in penicillin-binding proteins that lead to reduced binding to the beta lactam. The gram-positive pathogen Staphylococcus aureus produces beta lactamases, but beta lactamase inhibitors play a lesser role in treatment of these infections because the most resistant strains (methicillin-resistant Staphylococcus aureus) also use variant penicillin-binding proteins. β-lactamase inhibitors expand the useful spectrum of these β-lactam antibiotics by inhibiting the β-lactamase enzymes produced by bacteria to deactivate them.
•β-lactamase inhibitors with β-lactam core:
1. Tebipenem is the first carbapenem to be administered orally in the form of Tebipenem-Pivoxil. Structural and kinetic studies of tebipenem are available with M.tuberculosis beta-lactamase (BlaC). 6-Methylidene Penem2 is a newly designed beta-lactamase inhibitor and a very interesting one. After going inside the cell, when attacked by the enzyme beta-lactamase, it rearranges its molecular structure. In case of M. tuberculosis beta-lactamase, it exhibits 70 times higher activity than clavulanate. This rearrangement also makes it a good drug candidate to drug resistant beta-lactamases.
2. Boron based transition state inhibitors or BATSIs are very potent group of beta-lactamase inhibitors. A screen of a series of BATSIs against M. tuberculosis produces very interesting result. All the BATSIs with high inhibitory effects contain a benzoic carboxylic acid group. This is indeed a great break through in studying drug resistant beta-lactamases.
3. Clavulanic acid or clavulanate, usually combined with amoxicillin or ticarcillin
4. Sulbactam, usually combined with ampicillin or Cefoperazone
5. Tazobactam, usually combined with piperacillin
•Non-β-lactam β-lactamase inhibitors:
1. Avibactam, approved in combination with ceftazidime, currently undergoing clinical trials for combination with ceftaroline
2. Relebactam (previously known as MK-7655) is undergoing Phase III clinical trials as a treatment for pneumonia and bacterial infections.
______
Fighting antimicrobial resistance –The contribution of pharmacists:
Practice points for pharmacists:
•Reinforce directions for use.
•When there is concern that remaining antibiotics may be misused, consider dispensing the number of antibiotics required for the indicated duration of treatment, even if this means removing some tablets from a pack.
•Advise patients to dispose of unused antibiotics when the pack size is greater than needed for duration of therapy.
•Question people who return with a repeat prescription after a long period of time when it would be expected that the original infection would have resolved.
______
Reduce use of antibiotics in animals:
In 1997, European Union health ministers voted to ban avoparcin and four additional antibiotics used to promote animal growth in 1999. In 2006 a ban on the use of antibiotics in European feed, with the exception of two antibiotics in poultry feeds, became effective. In Scandinavia, there is evidence that the ban has led to a lower prevalence of antibiotic resistance in (nonhazardous) animal bacterial populations. As of 2004, several European countries established a decline of antimicrobial resistance in humans through limiting the usage antimicrobials in agriculture and food industries without jeopardizing animal health or economic cost. In 2000, the FDA announced their intention to revoke approval of fluoroquinolone use in poultry production because of substantial evidence linking it to the emergence of fluoroquinolone-resistant Campylobacter infections in humans. Legal challenges from the food animal and pharmaceutical industries delayed the final decision to do so until 2006. Fluroquinolones have been banned from extra-label use in food animals in the USA since 2007. However, they remain widely used in companion and exotic animals. Growing U.S. consumer concern about using antibiotics in animal feed has led to greater availability of “antibiotic-free” animal products. For example, chicken producer Perdue removed all human antibiotics from its feed and launched products labeled “antibiotic free” under the Harvestland brand in 2007. Consumer response was positive, and in 2014 Perdue also phased out ionophores from its hatchery and began using the “antibiotic free” labels on its Harvestland, Simply Smart, and Perfect Portions products.
______
______
New and old antibiotics:
_
New available antibiotics:
_
_
New antibiotics in clinical trials:
_
Technologies to facilitate Drug Discovery and Development:
One reason for the limited number of new antibiotics is that traditional sources of these products have been carefully evaluated to the point that virtually all promising antibacterial compounds have been identified. These sources include chemical libraries used by pharmaceutical companies and the small proportion of antibiotic-yielding bacteria and fungi that can be easily cultured. To improve this situation, investigators are developing new tools to identify novel sources of naturally occurring antibiotics, such as the iChip platform that facilitates screening of natural products from previously unculturable soil organisms by mimicking their native environment. Using this technology, researchers identified teixobactin, an antibiotic compound with a novel mechanism of action. Although teixobactin is still in the early stages of development, this experience suggests that the iChip technology could prove an effective way to identify new classes of antibiotics. Investigators are also exploring other untapped sources of products such as marine microbes and bacteria living in extreme conditions. Furthermore, antimicrobial peptides known as bacteriocins, produced by bacteria, are being evaluated for activity and feasibility as products. Although these new approaches to drug therapy hold promise, drug screening also could be improved with methods that better model physiologic conditions.
_
Teixobactin:
Eleftheria terrae (abbreviated E. terrae) is a recently discovered Gram-negative bacterium. E. terrae is a temporary name for the organism, as it was only discovered Fall 2014 and is undergoing scientific study. Teixobactin, a new antibiotic, was found in this bacterium making it an important discovery. The discovery of E. terrae could represent a new age of antibiotics, as teixobactin is the first new antibiotic discovered since the synthetic era of the 1980s. E. terrae´s production of teixobactin is prominent because recent tests have revealed that teixobactin binds differently than most normally used antibiotics which makes it harder for the bacteria being attacked to develop resistance. Experiments performed by Ling et al. have shown teixobactin is capable of binding to lipid precursors of peptidoglycan, which makes up part of bacterial cell walls. The results did not show any resistance to teixobactin in the organisms that were studied, including Staphylococcus aureus and Mycobacterium tuberculosis. These findings indicate that teixobactin’s target is not a protein, leading to the belief that the development of bacterial resistance to teixobactin is much less likely. However, several scientists caution that it is too early to conclude that teixobactin resistance would not develop in the clinical setting. Similar claims were made for vancomycin, yet resistance emerged soon after large-scale use in the 1980s. It is possible that genes encoding resistance to teixobactin are already present in soil bacteria. Resistance could also arise by mutation after prolonged use in patients.
_
Aspergillomarasmine A:
Antibiotic-resistant bacteria are disarmed with compound produced by Aspergillis fungus. It was harvested from a forest soil sample collected by one of Wright’s students who was hiking in Kejimkuijik National Park near Caledonia, N.S. The molecule is called AMA (aspergillomarasmine A). Once it was isolated, the researchers checked the chemical structure on a computer database and realized they had rediscovered a long-discarded compound, first identified in the 1960s as a cause of leaf wilt in plants. “It had been catalogued,” Wright said, “and then no one ever thought of it again as being useful until we repurified it one more time 50 years later.” AMA works not by killing the bacteria, but by turning off the resistance mechanism, making the bacteria vulnerable to an existing drug again. The goal is to develop AMA into a compound that could be used along with older antibiotics, a combination approach that, if it works in humans, could reverse antibiotic resistance, and breathe new life into the antibiotic arsenal.
_
Formicin:
Scientists at the APC Microbiome Institute at University College Cork have discovered a range of new antimicrobials that can kill many harmful bacteria. The latest antimicrobial added to their list of 20 new small proteins is called formicin. It was isolated from the intestine of a fish. The new antimicrobial, formicin was isolated from bacillus paralichenformis APC1576, a bacteria which was originally isolated from the intestine of a mackerel. Formicin can kill a wide range of harmful bacteria, including the Gram-positive pathogens, staphylococcus aureus, clostridium difficile, listeria monocytogenes and streptococcus mutans, a causative agent of tooth decay. Formicin is a member of a subclass of bacteriocins called lantibotics (as is) that contain modified amino acids that work together to kill the bacterial target. _
Lugdunin:
While the pharmaceutical companies are dithering about whether to invest in research on new antibiotics, researcher Andreas Peschel has authored a German study that discovered a promising candidate among the bacteria growing inside human noses. Mouth or gut microbes feast on a constant flow of food, while nasal bacteria dwell in a kind of wasteland, and as a result they have developed potent weapons to compete for scant nourishment and ensure their survival. The German researchers found a bacterium, Staphylococcus lugdunensis, that produces a chemical compound, possibly a powerful antibiotic against resistant bacterial. Tests in mice have shown that lugdunin is effective against a range of potentially deadly infections, including MRSA, which is acquired in hospitals and sickens 90,000 Americans annually.
_
XF-73 (Exeporfinium chloride) is an experimental drug candidate. It is an anti-microbial that works via weakening bacteria cell walls. It is a potential treatment for methicillin-resistant Staphylococcus aureus (MRSA) and possibly Clostridium difficile. It is being developed by Destiny Pharma Ltd. Structurally it is a dicationic porphyrin. It has completed a phase I clinical trial for nasal decolonisation of MRSA—being tested against 5 bacterial strains. It seems unlikely to cause MRSA to develop resistance to it. In 2014 a phase 1 clinical trial for nasal administration was run. As of February 2016 another phase 1 clinical trial (for nasal administration) is recruiting. Oral delafloxacin is an investigational anionic fluoroquinolone with a broad spectrum of antimicrobial activity, including activity against methicillin-resistant Staphylococcus aureus (MRSA) and other serious skin infections. The drug, which will likely receive priority review by FDA, could be approved by mid-2017.
_
Efflux Pump Inhibitors (EPI):
Recently, the use of efflux pump inhibitors has been investigated in order to improve and potentiate the activity of exported antibiotics. Such a strategy has been used to develop inhibitors that reduce the impact of efflux pumps on fluoroquinolone activity. As many efflux pumps possess significant structural homology, it is hoped that one inhibitor compound will be active against a range of pumps from different bacterial species. Most research has focused upon P. aeruginosa Mex efflux pumps and inhibitors of these. One such inhibitor lowered the MIC values of fluoroquinolones for both sensitive and resistant strains. In addition the frequency of selection of fluoroquinolone-resistant strains was also lower in the presence of the inhibitor, suggesting that efflux may be important in the selection of fluoroquinolone resistance. Similar observations have been made for S. pneumoniae and S. aureus. A requirement for an intact efflux system to allow the development of topoisomerase mutations and consequent fluoroquinolone resistance in E. coli has also been described. The link between active efflux and mutations in genes encoding the target site proteins suggests that the use of such inhibitors, in association with substrate antibiotics, may be useful by increasing both the activity and the range of species for which a drug may be effective. The design of new drugs and modification of existing molecules should also now be carried out with efflux pumps in mind. Structural alterations that reduce the ability of an antibiotic to be effluxed without compromising its activity may lead to the development of more potent compounds, certainly the ‘effluxability’ of drugs must now be considered, as agents are developed with regard to their overall efficacy and the likelihood of development of resistance. The combinational use of an EPI with antibacterial agents should potentiate the activity of antibacterials, and it would also reduce the frequency of emergence of resistant mutants. For example, the presence of the EPI Phe-Arg-β-naphthylamide resulted in a up to 2,000-fold reduction in the minimum inhibitory concentrations of antibacterials known to be substrates of the Campylobacter CmeABC pump, and the frequency of emergence of erythromycin-resistant mutants in C. jejuni was reduced more than 1,000 fold.
_
Scientists enhance ability of antibiotics to defeat resistant types of bacteria using molecules called PPMOs: a 2016 study:
Researchers at UT Southwestern Medical Center have developed a strategy to overcome a key defense that drug-resistant bacteria use to fend off antibiotic attack. Research teams created a synthetic compound that blocks a bacterial pump used to expel antibiotics. Bacteria that use these “efflux pumps” and that are treated with the new compound become sensitive to antibiotics they were previously resistant to. This is the first time researchers have tailored one of these synthetic compounds, part of a class called PPMOs or peptide-conjugated phosphorodiamidate morpholino oligomers, to target a specific efflux pump found in bacterial cell walls.. The PPMO compound itself doesn’t kill the bacteria but keeps it from expelling an antibiotic, allowing the drug to do its job. PPMOs are anti-sense molecules because they can mimic DNA or RNA and bind to mRNA’s genetic sequence like the other half of a zipper, blocking the bacterial machinery for building proteins. A PPMO designed to prevent creation of the AcrA protein significantly increased the efficacy of antibiotics against E. coli in cell models — from two- to 40-fold. The AcrA-PPMO also was effective against the human pathogens Klebsiella pneumoniae and Salmonella enterica, since those bacteria contain the same efflux pump with a matching gene sequence.
_
Laboratory experiments were combined with supercomputing modelling to identify molecules that boost the effect of antibiotics on bacteria: a 2016 study:
Researchers, including those from University of Oklahoma (OU) in the US, identified four new chemicals that seek out and disrupt bacterial proteins called “efflux pumps”, a major cause of antibiotic resistance in bacteria. The team focused on one efflux pump protein, known as AcrA, which connects two other proteins in a tunnel shape through the bacterial cell envelope. Disrupting this protein could essentially break the efflux pump – an approach unlike other drug design strategies that try to inhibit the biochemical processes. Using computational models produced by the Titan supercomputer, researchers screened various combinations of molecules and proteins to determine which ones were most disruptive to their formation. After more extensive analysis, they narrowed down our list to predict which molecules were most likely to disrupt the function of the efflux pump. Researchers then conducted laboratory experiments to confirm the disruption of the efflux pump and the antibiotic-reviving capability of four of the molecules selected.
_
Bezlotoxumab:
The US Food and Drug Administration (FDA) has approved a new drug for the treatment of Clostridium difficile infection (CDI). The drug bezlotoxumab is approved for the treatment of recurrent CDI in adults who are also receiving antibacterial drug therapy for CDI and are at high risk of CDI recurrence. It is a human clonal antibody that works by binding itself to a toxin produced by C difficile bacteria (toxin B) and neutralizing it. It should be used only in conjunction with antibacterial treatment for CDI. CDI, which can cause mild-to-severe diarrhea, is one of the most common healthcare-associated infections. According to the Centers for Disease Control and Prevention, CDI was responsible for nearly half a million illnesses in the United States in 2011, and 29,000 patients died within 30 days of infection. Recurrent CDI tends to affect patients over the age of 65 who have compromised immune systems.
_
Mannosides:
Fimbrion and GSK develop novel antibacterial therapy for Urinary Tract Infections which is a class of mannose-containing small molecule compounds called mannosides. Mannosides may be able to treat and prevent urinary tract infections (UTIs) without inducing antibiotic resistance. Options for treating bacterial infections, especially those caused by Gram-negative bacteria, are becoming increasingly limited due to the exponential increase in antibiotic resistance. Urinary tract infections are the third leading indication for antibiotic therapy and where multidrug-resistant bacteria are becoming increasingly common. Mannosides represent a new way of treating bacterial infections, by simply preventing the bacteria from being able to stick to the walls of the bladder thus allowing the body to naturally eliminate the infection.
_________
Block transfer of resistance genes: a 2012 study:
Researchers report they have found a way to disrupt the spread of antibiotic-resistance genes among S. pneumoniae bacteria, which can contribute to pneumonia, meningitis and other dangerous ailments. The bacterium Streptococcus pneumoniae — which can cause pneumonia, meningitis, bacteremia and sepsis — likes to share its antibiotic-defeating weaponry with its neighbors. Individual cells can pass resistance genes to one another through a process called horizontal gene transfer, or by “transformation,” the uptake of DNA from the environment. Researchers report that they can interrupt the cascade of cellular events that allows S. pneumoniae to swap or suck up DNA. The new findings, reported in the journal PLoS ONE, advance the effort to develop a reliable method for shutting down the spread of drug resistance in bacteria. Researchers focused on blocking a protein that, when it binds to a receptor in the bacterial cell membrane, spurs a series of events in the cell that makes the bacterium “competent” to receive new genetic material. The researchers hypothesized that interfering with this protein (called CSP) would hinder its ability to promote gene transfer.
________
Old antibiotics for MDR bacterial infections:
A new strategy to fight antimicrobial resistance: the revival of old antibiotics: a 2014 review:
The increasing prevalence of hospital and community-acquired infections caused by multidrug-resistant (MDR) bacterial pathogens is limiting the options for effective antibiotic therapy. Moreover, this alarming spread of antimicrobial resistance has not been paralleled by the development of novel antimicrobials. Resistance to the scarce new antibiotics is also emerging. In this context, the rational use of older antibiotics could represent an alternative to the treatment of MDR bacterial pathogens. It would help to optimize the armamentarium of antibiotics in the way to preserve new antibiotics and avoid the prescription of molecules known to favor the spread of resistance (i.e., quinolones). Furthermore, in a global economical perspective, this could represent a useful public health orientation knowing that several of these cheapest “forgotten” antibiotics are not available in many countries. This review aimed at providing a non-exhaustive collection of microbiological and clinical data on potentially useful older antibiotics. Authors considered the value of “forgotten” antibiotics for the treatment of (i) MDR Gram-negative bacterial infections (polymyxins, fosfomycin, mecillinam, temocillin, and nitrofurantoin); (ii) MDR Gram-positive infections [trimethoprim-sulfamethoxazole (TMP/SMX), tetracyclines, chloramphenicol, clindamycin, pristinamycin, rifampicin, and fusidic acid]; and (iii) MDR tuberculosis (clofazimine, amoxicilline-clavulanate, TMP/SMX, and minocycline). In summary, all old antibiotics included in this review have been associated with successful treatment of MDR bacterial infections. Their reuse represents a promising strategy to fight antimicrobial resistance. But some points remain critical for their revival in a sustainable manner and need further evaluations. First, clinical trials comparing the efficacy, safety, and cost effectiveness with current antibiotics are lacking. Second, prescription of old antibiotics needs also to be regulated by antibiotic stewardships and guided by resistance rates monitoring. Third, optimization of the usage of old antibiotics remains a priority that may be considered of similar importance to that of the assessment of new drugs.
_____
Antibiotics that ‘bomb’ microbes developed: a 2016 study:
Scientists have developed a new class of antibiotics using nanotechnology, which can be delivered at a particular or targeted location. The team from Indian Institute of Science, Bengaluru, and Bose Institute, Kolkata, said that analysis shows they act as “antimicrobial bombs”. Elaborating, they said these “targeted bombs” damage the bacterial membrane with increased power and strength as compared to current drugs, thus resulting in better efficiency. The researchers used a technique called the “nuclear magnetic resonance” technique, which helps in drawing a detailed 3-D picture of the combination. This enabled them to figure out the nature of the interaction between the nanoparticle and the antibiotic in the nano-drug combination at the atomic level for the first time. The antimicrobial molecules come close to the relatively large spherical nanoparticle, touch it momentarily and then move away like bees swarming around abeehive. The activity of the nanoparticle- peptide combination was found to be higher than the action of the individual components (nanoparticle and the peptide) alone, implying a synergistic action. According to the researchers, a large number of antimicrobial drug molecules can be packed with one nanoparticle, thereby achieving high density of these molecules. They can be delivered at a particular location, acting as an antimicrobial ‘bomb’. These bombs damage the bacterial membrane with increased potency as compared to the free non-conjugated drugs.
_____
_____
Leading Pharmaceutical Companies present Industry Roadmap to combat Antimicrobial Resistance:
Ahead of the United Nations General Assembly (UNGA) High-Level Meeting on Antimicrobial Resistance (AMR) in September 2016, 13 leading pharmaceutical companies presented a new roadmap that lays out four key commitments they will deliver by 2020 to reduce AMR. The commitments follow the principles identified and agreed upon in the Industry Declaration made at the 2016 World Economic Forum in Davos earlier this year, and reflect the companies’ intent to continue to proactively contribute to the global efforts to address AMR. This unprecedented collaboration between the pharmaceutical companies marks a major milestone in the fight against AMR. In presenting this roadmap, the signatory companies firmly demonstrate their shared ambition to overcome the staggering threat AMR represents for our society, economies, and citizens. They are committed to working to reduce the development of antimicrobial resistance, improve access to high-quality antibiotics, vaccines, and diagnostics, invest in R&D, and collaborate with governments and stakeholders to sustain those investments.
Specifically, this group of diversified companies commit to:
1. Reduce the environmental impact from the production of antibiotics, including a review of the companies’ manufacturing and supply chains, and work with stakeholders to establish a common framework for assessing and managing antibiotic discharge;
2. Help ensure antibiotics are used only by patients who need them, recognizing this requires concerted efforts from many stakeholders, through continued provider and patient education, an examination of the companies’ promotional activities, sharing of surveillance data with public health bodies and healthcare professionals, and collaboration with stakeholders to reduce uncontrolled antibiotic purchase;
3. Improve access to current and future antibiotics, vaccines, and diagnostics, including working with stakeholders to strengthen global health systems and address access bottlenecks; establishing new business models that balance access needs, appropriate antibiotic use, expanded vaccine coverage and adequate return to companies; and working to reduce the prevalence of substandard/counterfeit antibiotics in high-risk markets; and
4. Explore new opportunities for open collaborations between industry and the public sector to address challenges in the research and development of new antibiotics, vaccines, and diagnostics, recognizing the value these bring to society.
Signatory companies include:
Allergan
AstraZeneca
Cipla
DSM Sinochem
Roche
GSK
Johnson & Johnson
Merck
Novartis
Pfizer
Sanofi
Shionogi
Wockhardt
_____
_____
Probiotics:
The diverse communities of microbes that inhabit the human body (the microbiota) support human health in multiple ways and play crucial roles in protection against infectious diseases. The elements of the microbiota are diverse and include bacteria, fungi, and viruses. The potential to manipulate the microbiota to treat infection has already been demonstrated with the successful use of faecal transplant for treatment of C difficile infections. Use of this procedure to decolonize patients with multidrug-resistant organisms is the subject of active investigation. A non-chemical, non-classical approach to reversing the resistance problem would be the revival of the susceptible strains. By encouraging their regrowth and repopulation in areas where they have been severely reduced, they can replace resistant strains. One action is to re-introduce a susceptible, competitive flora. This approach has been exemplified by the use of probiotics. ‘Preempt’ is a commercial product consisting of different bacterial strains from adult hens, which, by exclusion colonization, prevents Salmonella colonization in chickens. Likewise, the GG lactobacillus fed to adults has shown efficacy in gastrointestinal illnesses without the need for antibiotics. Probiotics are the “friendly” bacterial flora living inside of our digestive tracts. Probiotics are defined as microorganisms that when administered in sufficient quantities may improve health. There are a variety of probiotics that have been studied for various health benefits. Their role in preventing drug-resistant infections in humans has not been clearly established.
_
Study explains how an Intestinal Microbe protects against more Dangerous Bacteria: a 2016 study:
Probiotics consisting of beneficial microorganisms, meanwhile, have the potential to deliver the benefits of antibiotics minus the pitfalls. Yet up until now, evidence of their efficacy has been largely anecdotal, their mechanisms of action poorly understood. Thanks to a pair of papers recently published in Science and Science Immunology by researchers at Rockefeller University, however, that is beginning to change. The studies demonstrate that an enzyme produced by a common intestinal microbe can protect the guts of worms and mammals alike from attack by harmful bacteria, and offer important insights into how it does so. Together, their findings could lead to the development of probiotics for use against such dangerous pathogens as Clostridium difficile, a leading cause of hospital-acquired infections. The researchers set out to investigate the probiotic potential of the microbe Enterococcus faecium in the roundworm Caenorhabditis elegans. Although E. faecium has long been used as a probiotic in livestock, its mode of action has never been clear. And it is far from being an ideal probiotic for use in humans: According to Kavita Rangan, first author of the Science paper and a member of Howard Hang’s Laboratory of Chemical Biology and Microbial Pathogenesis, E. faecium readily acquires antibiotic resistance in hospital settings and can lead to dangerous infections in people with compromised immune systems. Yet in a series of experiments, Rangan and her colleagues demonstrated that when fed E. faecium, C. elegans was better able to resist the harmful effects of infection by Salmonella typhimurium, an intestinal pathogen that in mammals invades the thin layer of epithelial cells lining the gut. “Salmonella was still able to colonize the intestine,” says Rangan, “but it didn’t cause the same tissue damage to the worms, and it didn’t kill them.” What’s more, they discovered that a particular enzyme called SagA, which is secreted in abundance by E. faecium, was sufficient to protect both worms and mice from Salmonella. And they showed that SagA worked its magic in mice even when produced by a different microbe called Lactobacillus plantarum–an entirely innocuous bug that is commonly used as a probiotic for human intestinal diseases, and which naturally inhabits environments ranging from sauerkraut to the human gut. In a series of complementary experiments, Virginia Pedicord–first author of the Science Immunology paper, and a postdoctoral fellow in both the Hang lab and Daniel Mucida’s Laboratory of Mucosal Immunology–and colleagues also showed that E. faecium protected mice against S. typhimurium. In addition, they demonstrated that E. faecium prevented the pathogen from passing through the epithelium and invading other organs such as the liver. Those experiments proved that E. faecium did not protect the mice by attacking S. typhimurium directly or by changing the balance of other microbes in the gut. “It doesn’t kill the bacteria, and it doesn’t deplete the microbiota, either,” Pedicord says. “It just prevents them from causing disease.” And it accomplishes this, she explains, by stimulating the production of specialized proteins that prevent pathogens from coming into contact with the epithelial layer in the first place–proteins that are generated by the epithelial cells themselves. The team confirmed that, as was the case with C. elegans, SagA was by itself sufficient to protect the mice from the ravages of S. typhimurium. And it also identified a clutch of receptors and antimicrobial peptides related to the innate immune system that must be present for the enzyme to do its work. But perhaps most strikingly, Pedicord and her colleagues showed that, when delivered by L. plantarum, SagA also protected the mice against C. difficile, a pathogen that causes debilitating and sometimes fatal gastroenteritis in human beings. As a result, the prospect of a benign probiotic that could defend against C. difficile while avoiding the problems associated with antibiotic treatment is welcome news.
________
Phage therapy:
Phage therapy is another method for treating antibiotic-resistant strains of bacteria. Phage therapy infects pathogenic bacteria with their own viruses, bacteriophages. Bacteriophages, also known simply as phages, infect and can kill bacteria. Phages insert their DNA into the bacterium, where it is transcribed and used to make new phages, after which the cell will lyse, releasing new phage able to infect and destroy further bacteria of the same strain. The high specificity of phage protects “good” bacteria from destruction. When applicable, bacteriophage therapy defeats antibiotic resistant bacteria. Phage therapy is the therapeutic use of lytic bacteriophages to treat pathogenic bacteria infections. Bacteriophage therapy is an important alternative to antibiotics. The success rate was 80–95% with few gastrointestinal or allergic side effects. British studies also demonstrated significant efficacy of phages against Escherichia coli, Acinetobacter spp., Pseudomonas spp and Staphylococcus aureus. Innovative phage-derived modalities also are being developed and evaluated. Lysins are lytic enzymes produced by phages that selectively destroy specific gram-positive pathogens with high specificity and disperse biofilms. The first commercially developed therapeutic product of this type, which targets S aureus, is currently in phase 1 clinical trials. Investigators are also using phages as a starting point to develop engineered products that can modulate bacterial cells, including their antibiotic-resistance mechanisms and virulence factors. Other innovative tools being explored for their ability to target resistant bacteria are systems that likely evolved to protect bacteria from phages: CRISPR (clustered regularly interspaced short palindromic repeats)–Cas systems, capable of precise genome editing. For instance, in enterococci, multidrug-resistant phenotypes seem to be correlated with the loss of functional CRISPR systems, suggesting that some bacteria may trade their CRISPR defense system for enhanced ability to acquire new resistance traits through horizontal gene transfer. Some investigators have proposed taking advantage of this observation by using phages to specifically deliver CRISPR systems to target resistance genes, ensuring that only resistant strains are targeted.
_____
Predatory Bacteria can cannibalize Drug-Resistant Bugs:
According to a new paper, these so-called “vampire” bacteria — named for their nasty habit of latching on to other microbes and sucking out their innards — have successfully hunted down and killed pneumonia inside the lungs of sick rats. The paper, authored by Daniel Kadouri of the Rutgers Medical School, presents compelling new evidence that vampire bacteria could become a viable weapon against the rising threat of superbugs. Predatory bacteria were first identified in 1962 as fast-swimming bacteria that attack and eat other bacteria. They’re found everywhere in the natural environment, in soils, oceans, rivers, lakes, and are even present in the human gut. The favorite strain of predator bacteria among researchers like Mitchell and Kadouri are called Bdellovibrio-and-like-organisms (BALOS). “These bacteria are able to penetrate the double membrane of Gram-negative bacteria, enter into the prey and consume it from the inside out,” says Mitchell. After sucking down a tasty meal, the BALOS use the energy boost to multiply. “A predator can produce anything from two to seven progeny from a single prey. Scientists don’t know enough about predatory bacteria to “program” the hungry suckers to target specific microbes. The best current technique, Mitchell explains, is to identify naturally occurring predators that have a “taste” for certain kinds of diseases. Once the predator bacteria are isolated in culture, researchers must feed them daily to keep them alive and reproducing. Then it’s time to set them loose on disease pathogens.
______
AMPs as potential antimicrobial therapy:
Antimicrobial peptides (AMPs) and the bacterial resistance mechanisms against them have been co-evolving for eons. A diverse array of life forms can produce AMPs, which can be used to promote immune defenses, nutrient acquisition or elimination of rival organisms from the environment. As a result, AMPs are found in a multitude of environments, ranging from mammalian tissues to soil and aquatic environments. This ubiquitous presence of AMPs in the environment provides strong selective pressure to drive the development of bacterial resistance against these peptides. AMPs are typically small, charged, amphipathic molecules that can be produced in a variety of structures. Though structurally diverse, most AMPs work by interacting with the bacterial cell surface, followed by disruption of cellular integrity. Accordingly, the majority of bacterial resistance mechanisms function by limiting the interaction of AMPs with the bacterial cell surface. Mechanisms of AMP resistance include trapping or sequestering of peptides, outright destruction of AMPs by proteolysis, removal of AMPs from the cell via active transport, and structural modification of the cell surface to avoid interaction with AMPs. Many of these resistance mechanisms are upregulated in response to AMPs, allowing the bacteria to adaptively counter the effects of AMPs. Loss of these resistance mechanisms can impair the ability of bacteria to colonize plant or animal hosts and can attenuate virulence for many pathogens. Mechanisms of resistance may evolve specifically within a bacterial lineage or be genetically transferred from other AMP-resistant organisms. At present, many AMPs are being investigated as potential antimicrobial therapies. AMP drug development should be carefully vetted because like any naturally-produced antimicrobial, cognate resistance mechanisms for AMPs are already present in the producer bacterium. While these resistance mechanisms may be found more frequently in producer strains, each has the propensity to be passed on to other genera or species within a shared environmental niche. Because the presence of AMPs provides high selective pressure for the acquisition of resistance, it is important to consider the potential for resistance mechanism transfer between bacteria when developing AMPs for clinical use. Additionally, depending on the AMP resistance mechanism that is selected for, a multitude of issues may arise if the mechanism of resistance is broad-spectrum. A broad-spectrum AMP resistance mechanism could restrict the already limited clinical treatment options for use against some Gram-positive pathogens and may undermine our own immune response by conferring resistance to our own innate immune system peptides.
_
Engineers design AMPs against bacteria: a 2016 study:
Antimicrobial peptides, produced by all living organisms as part of their immune defenses, kill microbes in several different ways. First, they poke holes in the invaders’ cell membranes. Once inside, they can disrupt several cellular targets, including DNA, RNA, and proteins. These peptides also have another critical ability that sets them apart from traditional antibiotics: They can recruit the host’s immune system, summoning cells called leukocytes that secrete chemicals that help kill the invading microbes. Scientists have been working for several years to try to adapt these peptides as alternatives to antibiotics, as bacteria become resistant to existing drugs. Naturally occurring peptides can be composed of 20 different amino acids, so there is a great deal of possible variation in their sequences. A team of researchers at MIT, the University of Brasilia, and the University of British Columbia has now engineered an antimicrobial peptide that can destroy many types of bacteria, including some that are resistant to most antibiotics. In this study, the researchers began with a naturally occurring antimicrobial peptide called clavanin-A, which was originally isolated from a marine animal known as a tunicate. The original form of the peptide kills many types of bacteria, but the researchers decided to try to engineer it to make it even more effective. Antimicrobial peptides have a positively charged region that allows them to poke through bacterial cell membranes, and a hydrophobic stretch that enables interaction with and translocation into membranes. The researchers decided to add a sequence of five amino acids that would make the peptides even more hydrophobic, in hopes that it would improve their killing ability. This new peptide, which they called clavanin-MO, was very potent against many bacterial strains. In tests in mice, the researchers found that it could kill strains of Escherichia coli and Staphylococcus aureus that are resistant to most antibiotics. By using the thereby designed molecule, infected animals could be rescued from infections that were untreatable (and thus lethal) with standard antibiotic therapies. Whether or not this specific compound makes it to the hospital in the short run, the strategy of merging a killer activity with immunomodulant properties opens new avenues for dealing with the phenomenal AMR problem. The researchers also found that these peptides can destroy certain biofilms, which are thin layers of bacterial cells that form on surfaces. That raises the possibility of using them to treat infections caused by biofilms, such as the Pseudomonas aeruginosa infections that often affect the lungs of cystic fibrosis patients. Or, they could be embedded into surfaces such as table tops to make them resistant to microbial growth. Other possible applications for these peptides include antimicrobial coatings for catheters, or ointments that could be used to treat skin infections caused by Staphylococcus aureus or other bacteria. If these peptides are developed for therapeutic use, the researchers anticipate that they could be used either in stand-alone therapy or together with traditional antibiotics, which would make it more difficult for bacteria to evolve drug resistance. The researchers are now investigating what makes the engineered peptides more effective than the naturally occurring ones, with hopes of making them even better.
_
Devil’s milk contains AMPs that kill superbugs: a 2016 study:
Tasmanian devils are the world’s largest carnivorous marsupials and among Australia’s most beloved creatures. Devil’s milk has proved to be an unlikely weapon in the increasingly desperate global fight against superbugs. Australian researchers have discovered that peptides contained in the milk of Tasmanian devils can kill some of the most deadly bacterial and fungal infections, including golden staph. The devil milk peptides hail from a family of antimicrobials called cathelicidins, which act as natural antibiotics. All animals have them. Devils have six varieties, while humans have just one. Having scanned the devil’s genome and discovered the six naturally occurring antimicrobial peptides, researchers from Sydney University set about replicating them artificially. They then tested the peptide’s effectiveness at killing some of the most harmful bacteria known to humans. Among the drug resistant bacteria the devil peptides killed was golden staph, or methicillin-resistant Staphylococcus aureus.
_
Researchers Nanoengineer Kryptonite for Antibiotic Resistant Superbugs: a 2016 study:
A research team at the University of Melbourne led by graduate student Shu Lam has nanoengineered a peptide polymer that attacks and kills a deadly class of drug-resistant bacteria. The polymer selectively targets bacteria and is not harmful to healthy human tissue. A superbug that is resistant to a last-line-of-defense antibiotic failed to develop resistance to the polymer after many generations of exposure. The team at the University of Melbourne thought outside the antibiotic box and explored antimicrobial peptides as a way to combat AMR. Antibiotics poison bacteria; antimicrobial peptides tear them apart. Lam and her team built star-shaped “structurally nanoengineered antimicrobial peptide polymers” (SNAPPs) and used them to attack drug-resistant bacteria both in vitro and in vivo. They reported their results in the journal Nature Microbiology. For the in vitro tests, four bacterial pathogens and two colistin-resistant superbugs were exposed to SNAPPs. Exposure took place in both a nutrient-rich bacterial growth medium and a minimum essential growth medium which is the standard medium for mammalian cells. The SNAPPs were highly effective at killing all six strains of bacteria in both mediums.
_______
Know your enemy, bacteria or virus:
This suggests one way to reduce the development of resistance would be to have diagnostic kits that ruled bacterial infection in or out on the spot. Such kits would have to be very quick, cheap and convenient indeed to supplant pre-emptive antibiotics.
So how do you know if you have a viral or bacterial infection?
1. Procalcitonin level:
Identifying whether the cause of inflammation in patients is of bacterial origin has been an important area of development in the clinical laboratory. Several clinical laboratory tests have been applied to the diagnosis of sepsis. The broth culture method is the gold standard for the diagnosis of bacterial infection, but a definitive result can take 24 hours or more before a conclusive diagnosis. A number of the inflammatory markers, such as leukocyte cell count, C reactive protein (CRP), and cytokines (TNF-α, IL-1β, or IL-6), have been applied in the diagnosis of inflammation and infection, but their lack of specificity has generated a continued interest to develop more specific clinical laboratory tests. One promising marker has been procalcitonin (PCT), whose concentration has been found to be elevated in sepsis. Owing its specificity to bacterial infections, PCT has been proposed as a pertinent marker in the rapid diagnosis of bacterial infection, especially for use in hospital emergency departments and intensive care units. A growing body of evidence supports the use of procalcitonin (PCT) to improve diagnosis of bacterial infections and to guide antibiotic therapy. For patients with upper and lower respiratory tract infection, post-operative infections and for severe sepsis patients in the intensive care unit, randomized-controlled trials have shown a benefit of using PCT algorithms to guide decisions about initiation and/or discontinuation of antibiotic therapy.
2. Adopt Rapid Diagnostic Tests:
Molecular methods are coming fast. We now have a polymerase chain reaction test for the detection of MRSA, vancomycin-resistant Enterococcus, Neisseria gonorrhoeae, Chlamydia trachomatis, group B Streptococcus, tuberculosis, Candida albicans, and many others. Coming soon are tests that will detect practically every bacterium as well as other pathogens, making an etiologic diagnosis to facilitate antibiotic decision-making within 1-2 hours of collecting the culture. Interpretation will be tricky, however, because many specimens will need quantitation and there will be a predictable need for substantial stewardship.
3. New diagnostic tool could help doctors make more informed prescriptions.
The diagnostic tool uses genetically engineered bacteria to detect the presence of a bacterial infection in a patient’s blood sample. It can distinguish between a viral and bacterial infection by detecting a protein known as lipocalin. This protein is produced in high levels by cells of the immune system in response to bacterial infections. The protein’s function is to bind to small molecules which bacteria use to access iron in order to grow. The Sheffield team, which consists of students from a range of science, engineering and medicine disciplines, are developing the device so that genetically modified bacteria mixes with a patient’s blood sample and turn florescent when there are low levels of the lipocalin protein – indicating a viral not bacterial infection. The main aim behind this project is to create more informed prescriptions to address the ever increasing resistance against antibiotics that we face today. Antibiotic resistance is a huge problem and this is why students chose to base this project on it. We may not be able to reverse it, but with our device, we could potentially slow it down. What’s even more interesting is that using bacteria to detect bacteria.
4. Can you differentiate bacterial from viral infections based on the CBC?
No—the complete blood count (CBC) alone does not have adequate sensitivity or specificity to tell bacterial from viral infections. The bacterial infections increase neutrophil counts while viruses increase lymphocyte count. For acutely febrile patients, the presence of an elevated white blood cell (WBC) count with elevated band forms has dogmatically been thought of as a marker for bacterial infection. Current literature, however, does not support this. The CBC cannot be used in isolation to differentiate bacterial from viral illness. The CBC can, however, augment clinical data from the history and physical examination to predict the likelihood of serious bacterial illness. As a result, numerous diagnostic criteria, each incorporating elements of the CBC, have been developed in an attempt to accurately differentiate bacterial from viral illness in acutely febrile patients.
WBC markers:
How good are they at predicting serious bacterial infection?
| VARIABLE | CUTOFF | SENSITIVITY | SPECIFICITY | LR (95% CI) |
| White blood cell count | 15,000/mm3 | 64%–82% | 67%–75% | 1.9–2.7 (1.1–3.8) |
| Absolute neutrophil count | 10,000/mm3 | 64%–76% | 76%–81% | 3.0–3.3 (1.6–6.2) |
| LR, likelihood ratio; CI, confidence interval. | ||||
_______
Antivirulence Strategies:
Factors that contribute to pathogen virulence—such as toxins, iron acquisition systems, secretion systems, quorum-sensing pathways, adhesins, and biofilm formation—have the potential to be exploited as novel therapeutic targets. Selectively targeting virulence factors is attractive because this strategy does not affect microbe viability and thus does not exert the selective pressure associated with conventional antimicrobials. The goal of antivirulence therapeutics is to reduce pathogenicity while allowing the host to clear the bacterial infection. This approach has the added benefit of preserving the host microbiota. However, new preclinical testing approaches and models will need to be developed if antivirulence therapeutics are to be advanced to the clinic. For example, animal efficacy models will need to more accurately reflect clinical progression of disease and new in vitro assays will need to be developed.
_______
Miscellaneous measures to control AMR:
How honey kills bacteria: a 2010 study:
With the rise in prevalence of antibiotic-resistant bacteria, honey is increasingly valued for its antibacterial activity. To characterize all bactericidal factors in a medical-grade honey, authors used a novel approach of successive neutralization of individual honey bactericidal factors. All bacteria tested, including Bacillus subtilis, methicillin-resistant Staphylococcus aureus, extended-spectrum β-lactamase producing Escherichia coli, ciprofloxacin-resistant Pseudomonas aeruginosa, and vancomycin-resistant Enterococcus faecium, were killed by 10–20% (v/v) honey, whereas ≥40% (v/v) of a honey-equivalent sugar solution was required for similar activity. Honey accumulated up to 5.62 ± 0.54 mM H2O2 and contained 0.25 ± 0.01 mM methylglyoxal (MGO). After enzymatic neutralization of these two compounds, honey retained substantial activity. Using B. subtilis for activity-guided isolation of the additional antimicrobial factors, authors discovered bee defensin-1 in honey. After combined neutralization of H2O2, MGO, and bee defensin-1, 20% honey had only minimal activity left, and subsequent adjustment of the pH of this honey from 3.3 to 7.0 reduced the activity to that of sugar alone. Activity against all other bacteria tested depended on sugar, H2O2, MGO, and bee defensin-1. Thus, authors fully characterized the antibacterial activity of medical-grade honey.
_
Maggots kill antibiotic resistant bacteria:
Nowadays, it is generally recognized that maggots can excrete a complex mixture of substances into a wound that can clean the wound and stimulate its healing. One aspect of the complex mode of action of this effect is the antibacterial activity. Despite several reports concerning such antibacterial properties (Greenberg, 1968; Pavillard & Wright, 1957; Simmons, 1935; Thomas et al., 1999) and some attempts to determine what these antibacterial compounds are (Bexfield et al., 2004), the main components of this effective antimicrobial activity are still not known. However, it can be expected that several different antibacterial components, like oligopeptides (Bexfield et al., 2004), disinfectants (Erdmann & Khalil, 1986) and low pH (Baer, 1931), act synergistically, as was proposed for Phormia terraenovae (Pavillard & Wright, 1957). Moreover, some of these antibacterial components could also be synthesized by some of the maggots’ gut commensals, as proposed by Greenberg (1968). Proteus mirabilis is one such commensal, indicating the general occurrence of members of the genus Proteus in flies of the family Calliphoridae. Therefore, if some bacteria present in the gut of the maggots survive the complex mode of maggot antibacterial action, some antibacterial selectivity of the excreta/secreta mix of substances would be expected. Since multiple factors are responsible for the antibacterial activity of maggot excretions, it is evident that the efficiency of the excreta/secreta, and hence of larval therapy, depends on one hand on the ‘quality’ of the maggots, and on the other hand on the microbiological and physico-chemical conditions present in their immediate environment.
Maggots rid patients of antibiotic-resistant infection, MRSA: a 2007 study:
Medical researchers are ridding diabetic patients of the superbug MRSA — by treating their foot ulcers with maggots. The scientists used green bottle fly larvae to treat 13 diabetic patients whose foot ulcers were contaminated with MRSA and found all but one were cured within a mean period of three weeks, much quicker than the 28-week duration for the conventional treatment. This is very exciting. Authors have demonstrated for the first time the potential of larval therapy to eliminate MRSA infection of diabetic foot ulcers. If confirmed in a randomized controlled trial, larval treatment would offer the first non-invasive and risk-free treatment of this increasing problem and a safe and cost-effective treatment in contrast to the expensive and potentially toxic antibiotic remedies.
__
New sources of antibiotics:
Since Alexander Fleming’s serendipitous 1928 observation that a Penicillium fungus prevented growth of staphylococci bacteria, the search for new antibiotics has largely been focused on fungi and microbes living in the soil, in the hopes of discovering another natural product with the broad effectiveness and low toxicity of penicillin. But as more recent searches result in disappointment, some investigators are turning to new sources—plants, insects, and marine organisms—to find antibiotics that can kill our most common and persistent pathogens. When chemist Simon Gibbons of the University College London School of Pharmacy went in search of plants harboring compounds with antimicrobial properties in 2008, he paid particular attention to those that have been used in traditional medicine—especially for wound healing. “If a plant is used as a wound-healing agent, it’s quite likely that it contains chemicals that kill the bacteria in the wound,” he says. Although best known for its psychoactive properties, cannabis, historically ingested in parts of Afghanistan and India to treat infection, fit the bill. Gibbons and his colleagues isolated five cannabinoids from Cannabis sativa and found that each one was effective against MRSA (J Nat Prod, 71:1427-30, 2008). “It hasn’t been confirmed in vivo, but certainly in the lab, we know that these things kill drug-resistant bacteria,” he says. Gibbons has also found chemicals with antibiotic properties in other familiar plant groups. For instance, plants in the Allium genus, which includes garlic and onions, produce sulfur-containing compounds that have activity against MRSA and Mycobacterium (J Nat Prod, 72:360-65, 2009). And many of the hypericums—the family that includes St. John’s wort—make chemicals called acylphloroglucinols that also effectively kill MRSA in vitro (J Nat Prod, 75:336-43, 2012). “We’ve had leads . . . and a series of compounds, which have been patented,” says Gibbons, and those compounds are now being synthesized and modified to improve their activity. The bright yellow spice that gives curries their rich color and smoky flavor, turmeric is far more than a culinary tool—it has amazing healing properties. MRSA typically forms in wounds or boils. Turmeric acts as an antibacterial agent and can be taken internally or applied directly to the skin.
_______
Public education campaign:
Public education campaigns have exerted a positive influence on inappropriate prescribing in countries such as Belgium and France. For example, a national media campaign in Belgium coincided with a 36% reduction in antibiotic prescriptions over 7 years (although other factors likely contributed as well). Other countries, including the United States, have launched education campaigns (e.g., the Centers for Disease Control and Prevention’s “Get Smart About Antibiotics Week” or the “Medicines With the Red Line” public awareness campaign in India). Importantly, these programs are still relatively new and conclusive evidence of their effectiveness is not yet available. Public campaigns can work but they are costly. France conducted a national campaign to convince patients and providers to do better, with a target of a 25% reduction in antibiotic prescriptions in the entire country. They achieved a 26% reduction! Efforts targeting primary care clinicians also can change practice: a recent randomized clinical trial of behavioral interventions (e.g., lists comparing levels of inappropriate prescribing among peers) demonstrated statistically significant decreases in inappropriate prescribing. Similarly, in a cluster randomized trial in which investigators offered an educational module and personalized feedback designed to reduce broad-spectrum antibiotic prescriptions for acute respiratory tract infections in children, intervention sites demonstrated a 12.5% decline in prescriptions for broad-spectrum agents compared with a 5.8% decline in control settings.
_
Antibiotic reduction campaigns do not necessarily reduce resistance: a 2013 study:
Antibiotic use — and misuse — is the main driver for selection of antibiotic resistant bacteria. This has led many countries to implement interventions designed to reduce overall antibiotic consumption. Now, using methicillin resistant Staphylococcus aureus as an example, scientists warn that simply reducing antibiotics consumption does not necessarily reduce resistance. The success of antibiotic reduction programs depends on which antibiotics are reduced, because some select more strongly for resistance than others. For instance, in the case of S. aureus, reducing use of clindamycin and methicillin lead to decreased resistance, while reducing use of penicillins does not, since most S. aureus, including MRSA, are already resistant to penicillin, explains Temime. Additionally, efforts to reduce antibiotic use must be coordinated between hospitals and the community, since either can feed resistant bacteria into the other, undermining reduction efforts, says Temime. In 2002-2003, a national program reduced antibiotic use in France by 10 percent. However, it fell short of the full potential for reducing resistance because it failed to target those antibiotics that generate the most resistance, says Temime. She and her collaborators developed a mathematical model of MRSA circulation, which correctly simulated that reduction, post-facto. They then performed a number of simulations of reductions in antibiotic use, which demonstrated the complexities of reduction efforts. “We found that the reduction in MRSA hospital rates could have been much larger than it actually was following the 2002 antibiotic reduction campaign,” says Temime. “Our results also suggest that changes in the distribution of antibiotics prescribed for non-hospitalized patients actually limited the impact of the antibiotic reduction campaign in French hospitals.” Their research shows that class-specific changes in antibiotic use, rather than overall reductions, need to be considered in order to achieve the greatest benefit from antibiotic reduction campaigns, says Temime. “This underlines the importance of generating surveillance data on both antibiotic class-specific changes in antibiotic use and antibiotic resistance in the years following an antibiotic reduction campaign. We believe that this research may help health policy makers and physicians in the design of more efficient antibiotic reduction campaigns.”
_______
_______
ARDB—Antibiotic Resistance Genes Database: a 2008 venture:
Several mechanisms have been characterized through which bacteria become resistant to antibiotics: (i) the production of enzymes that digest/metabolize the antibiotic; (ii) efflux pumps that eliminate the drug from the cell; (iii) modifications to the cellular target of the antibiotic that prevent binding; (iv) activation of an alternate pathway that bypasses drug action; and (v) particularly for gram-negative bacteria, down-regulation or elimination of transmembrane porins through which drugs enter the cell. The annotation information commonly associated with genes deposited in public databases is insufficiently detailed for representing this variety of resistance mechanisms and the additional meta-information relevant in this context. Specifically, each resistance gene is associated with a resistance profile (set of antibiotics or classes of antibiotics targeted by the gene), yet this information is usually not available. Second, resistance often requires the cooperation of multiple genes, usually within a same operon [e.g. vancomycin resistance VanA operon requires seven genes], while most annotation information is targeted at individual genes. Finally, resistance frequently results from modifications to, or the disruption of an individual gene (e.g. modifications of the drug target), information incompatible with standard annotation procedures. Consequently, specialized resources are necessary for annotating and cataloging information related to antibiotic resistance. Several recent efforts have been made to partially unify this information, such as Antibiotic Resistance Genes Online (ARGO), MvirDB and a compendium of TEM β-lactamase genes at the Lahey Clinic. All, however, have limited functionality. ARGO only contains part of β-lactamase, vancomycin and tetracycline resistance genes. In addition, it does not include rich annotation information such as resistance profile, mechanism of action, operon information or gene sequence. Furthermore, many of the links between ARGO and GenBank target incorrect records (e.g. links to a genome instead of the relevant gene record). MvirDB is a broad repository of virulence-associated genes, including toxins, virulence factors and antibiotic resistance. The latter information is simply a replicate of the ARGO database. The Lahey Clinic website is a comprehensive collection of TEM type β-lactamases, which attempts to standardize the nomenclature for these genes. In addition to these specialized resources, antibiotic resistance information can be extracted in a restricted manner from GenBank and SwissProt, databases that lack many important types of information relevant in this domain.
_
To address the limitations of currently available public resources, and to facilitate the identification and characterization of antibiotic resistance genes, authors have created a manually curated database [Antibiotic Resistance Genes Database (ARDB)] unifying most of the publicly available genes and related information. Their motivations in creating ARDB are (i) to provide a centralized compendium of information on antibiotic resistance; (ii) to facilitate the consistent annotation of resistance information in newly sequenced organisms; and (iii) to facilitate the identification and characterization of new genes. They believe this resource will be found useful by a broad range of scientists, including microbiologists, clinicians and the bio-defense research community. The treatment of infections is increasingly compromised by the ability of bacteria to develop resistance to antibiotics through mutations or through the acquisition of resistance genes. Antibiotic resistance genes also have the potential to be used for bio-terror purposes through genetically modified organisms. In order to facilitate the identification and characterization of these genes, they have created a manually curated database—the Antibiotic Resistance Genes Database (ARDB)—unifying most of the publicly available information on antibiotic resistance. Each gene and resistance type is annotated with rich information, including resistance profile, mechanism of action, ontology, COG and CDD annotations, as well as external links to sequence and protein databases. This database also supports sequence similarity searches and implements an initial version of a tool for characterizing common mutations that confer antibiotic resistance. The information they provide can be used as compendium of antibiotic resistance factors as well as to identify the resistance genes of newly sequenced genes, genomes, or metagenomes. Currently, ARDB contains resistance information for 13,293 genes, 377 types, 257 antibiotics, 632 genomes, 933 species and 124 genera. ARDB is available at http://ardb.cbcb.umd.edu/. ARDB unifies most of the publicly available antibiotic resistance genes and provides a reliable annotation service to researchers investigating the molecular basis for resistance in bacteria. Because of the large diversity and the rapid identification of new resistance genes, the current version of ARDB is just a catalog of currently available information, and will continue to be updated over the coming months and years.
______
______
The Regulatory Side of Resistance:
Regulations are the responsibility of governments, and for some of us, that clearly indicate that something inevitably is going to be completely wrong, as governments seldom do something right. However, on the one hand, some issues must be regulated regarding antibiotic usage, production, and marketing; on the other hand, we pay handsomely to governments and it is only fair to expect them to do their work every once in a while. The main question is, what should be regulated? In the so-called developing countries, one of the first things that must be regulated is the sale of antibiotics: antibiotics, as most other drugs, can be purchased without medical prescription, on over-the-counter basis. This leads to self-prescription, which we can make it inclusive of all non-medically prescribed drugs, often from the advice of a relative, a friend, or even the sales clerk at the drug store, all of whom lack any kind of medical training. It is difficult to assess how much antibiotics are sold and used in this way. As an example, a survey in Mexico City revealed that at selected drug stores, about 30% of antibiotics were sold without medical prescription (Ama´ bile-Cuevas et al. 1998). It is much more difficult to assess the burden this practice poses upon resistance, but we can assume that self-prescription would be wrong more often than medical one, leading to inadequate choice of drugs, inadequate doses and/or length of treatments, and, of course, a more common use of antibiotics where they were not necessary. A simple, draconian approach to this problem would be simple: to ban the sale of antibiotics without medical prescription. This has been made in a few Latin American countries, which we can only hope would attempt to record the impact of the measure. However, a trait of the ‘‘developing’’ countries is that a huge fraction of their populations lack access to medical care; for them, self-prescription might very well be the only means of access to antibiotics. Since we do not know how many lives are actually being saved by self-prescribed antibiotics, banning the free sale of such drugs could prevent a potential jeopardy (rising resistance) by causing an actual damage, denying access to life-saving drugs. So we must wait to see the impact of a prohibition, before extending it to all countries. Besides, and based only on the Mexican results cited before, medical prescription still accounts for more antibiotic abuse than self-prescription: if 70% of antibiotics are prescribed by a physician, and such prescriptions are 50% wrong, then this makes for 35% of all antibiotics sold at drugstores, more than the 30% accountable by self-prescription. The mere suggestion of the regulation of the clinical use of antibiotics almost always results in the angry opposition of clinicians: they definitely know better than a simple rule, and are trained to understand signs and symptoms, and to decide when, where and which drug to use. Unfortunately, that is simply not true. Clinicians often use antibiotics to treat viral infections or even ailments that are not infectious at all; have difficulties to cope with the variety of microorganisms, drugs, mechanisms of action, pharmacokinetics and bioavailabilities, and resistance trends that must be considered before selecting a drug; and do not know what to expect from and what to do with a clinical microbiology report. For those who acknowledge some or all of these problems, the solution should lie upon educational efforts, perhaps only aided by some of the ubiquitous ‘‘guidelines’’ that pharmaceutical companies are only too kind to support; regulation, they say, hinders good medical practice. Although, as mentioned before, education should and must be improved, some basic rules must be set up and enforced. For those physicians that do not adhere to these rules, legal consequences should be established; we are well beyond the point where just admonishing would do. These rules should include, but not be limited to, the use of antibiotics when not needed, as in non-infectious, viral, or self-limited diseases; the reckless use of wide-spectrum antibiotics; the unwarranted use of antimicrobial prophylaxis; and errors in dosing. Another important issue calling for stronger regulatory efforts is drug production and marketing. Although this is important for all drugs, it is particularly relevant for antibiotics, as low-quality antimicrobials do not ‘‘only’’ affect the patient, but can also include effects upon microbial populations. Final issues regarding antibiotic usage from the regulatory point of view include their non-clinical use, namely for agricultural purposes. Also, strict surveillance upon marketing strategies from pharmaceutical companies is necessary to prevent them to push for inadequate uses of their products.
_
Legal framework:
Some global health scholars have argued that a global, legal framework is needed to prevent and control antimicrobial resistance. For instance, binding global policies could be used to create antimicrobial use standards, regulate antibiotic marketing, and strengthen global surveillance systems. Ensuring compliance of involved parties is a challenge. Global antimicrobial resistance policies could take lessons from the environmental sector by adopting strategies that have made international environmental agreements successful in the past such as: sanctions for non-compliance, assistance for implementation, majority vote decision-making rules, an independent scientific panel, and specific commitments.
_
National Antibiotic policy:
The CDDEP report stresses that antibiotic stewardship – reducing the inappropriate and unnecessary use of antibiotics – is key to controlling antibiotic resistance. It lays out six strategies that should be incorporated in national antibiotic policies to halt its spread:
(i) reduce antibiotic need through improved water, sanitation and immunisation;
(ii) improve hospital infection control;
(iii) disincentivise antibiotic overuse/misuse and incentivise antibiotic stewardship;
(iv) reduce and phase out subtherapeutic antibiotic use in agriculture;
(v) educate health professionals, policy makers and the public on sustainable antibiotic use; and
(vi) ensure political commitment to meet the threat of antibiotic resistance.
The report advises that limiting overuse and misuse of antibiotics are the only sustainable solutions. A rampant rise in antibiotic use poses a major threat to public health. We need to focus 80% of our global resources on stewardship and no more than 20% on drug development. No matter how many new drugs come out, if we continue to misuse them they might as well have never been discovered.
_
In 2014, the WHO stated:
1. People can help tackle resistance by:
◦using antibiotics only when prescribed by a doctor;
◦completing the full prescription, even if they feel better;
◦never sharing antibiotics with others or using leftover prescriptions.
2. Health workers and pharmacists can help tackle resistance by:
◦enhancing infection prevention and control;
◦only prescribing and dispensing antibiotics when they are truly needed;
◦prescribing and dispensing the right antibiotic(s) to treat the illness.
3. Policymakers can help tackle resistance by:
◦strengthening resistance tracking and laboratory capacity;
◦regulating and promoting appropriate use of medicines.
4. Policymakers and industry can help tackle resistance by:
◦fostering innovation and research and development of new tools;
◦promoting cooperation and information sharing among all stakeholders.
_
To combat the threat to human health and biosecurity from antimicrobial resistance, an understanding of its mechanisms and drivers is needed. Emergence of antimicrobial resistance in microorganisms is a natural phenomenon; yet antimicrobial resistance selection has been driven by antimicrobial exposure in health care, agriculture, and the environment. Onward transmission is affected by standards of infection control, sanitation, access to clean water, access to assured quality antimicrobials and diagnostics, travel, and migration. Strategies to reduce antimicrobial resistance by removing antimicrobial selective pressure alone rely upon resistance imparting a fitness cost, an effect not always apparent. Minimising resistance should therefore be considered comprehensively, by resistance mechanism, microorganism, antimicrobial drug, host, and context; parallel to new drug discovery, broad ranging, multidisciplinary research is needed across these five levels, interlinked across the health-care, agriculture, and environment sectors. Intelligent, integrated approaches, mindful of potential unintended results, are needed to ensure sustained, worldwide access to effective antimicrobials.
_
Detecting, preventing, and controlling antibiotic resistance requires a strategic, coordinated, and sustained effort. It also depends on the engagement of governments, academia, industry, healthcare providers, the general public, and the agricultural community, as well as international partners. Success in this effort will require significant efforts to: minimize the emergence of antibiotic-resistant bacteria; preserve the efficacy of new and existing antibacterial drugs; advance research to develop improved methods for combating antibiotic resistance and conducting antibiotic stewardship; strengthen surveillance efforts in public health and agriculture; develop and promote the use of new, rapid diagnostic technologies; accelerate scientific research and facilitate the development of new antibacterial drugs, vaccines, diagnostics, and other novel therapeutics; maximize the dissemination of the most up-to-date information on the appropriate and proper use of antibiotics to the general public and healthcare providers; work with the pharmaceutical industry to include information on the proper use of over-the-counter and prescription antibiotic medications for humans and animals; and improve international collaboration and capabilities for prevention, surveillance, stewardship, basic research, and drug and diagnostics development.
________
________
Applications of antibiotic resistance:
Antibiotic resistance is an important tool for genetic engineering. By constructing a plasmid that contains an antibiotic-resistance gene as well as the gene being engineered or expressed, a researcher can ensure that, when bacteria replicate, only the copies that carry the plasmid survive. This ensures that the gene being manipulated passes along when the bacteria replicates.
In general, the most commonly used antibiotics in genetic engineering are “older” antibiotics. These include:
•ampicillin
•kanamycin
•tetracycline
•chloramphenicol
In industry, the use of antibiotic resistance is disfavored, since maintaining bacterial cultures would require feeding them large quantities of antibiotics. Instead, the use of auxotrophic bacterial strains (and function-replacement plasmids) is preferred.
______
______
Discussion:
_
Lessons learned about Antibiotic Resistance:
1. Given enough antibiotic and time, resistance will appear. For example, the penicillin-resistant Streptococcus pneumoniae took 25 years to become a clinical problem; fluoroquinolone-resistant Enterobacteriaceae took 10 years to emerge clinically.
2. Resistance is progressive, moving from low to intermediate to high levels.
3. Organisms that are resistant to one antibiotic will likely become resistant to others. For example, tetracycline resistance in Neisseria gonorrhoeae first appeared among strains with existing resistance to penicillin.
4. Once selected, drug resistance will not disappear, although it may decline slowly. This gradual decrease in resistance is associated with poorly reversible genetic and environmental factors. No counterselective steps against resistant bacteria now exist.
5. When antibiotics are used by any patient, this use affects other people by changing the microbiology in both the immediate and the extended environment.
_
We began the antibiotic era with a full-fledged attack on bacteria. It was a battle misconceived and one in which we cannot be the winner. We cannot destroy the microbial world in which we have evolved. The best solution now is to take a broader view of the microbial world. While focusing on the pathogens, our efforts should act in ways that impact fewer commensal flora. We need to forget ‘overcome and conquer’ and substitute ‘peace’ when regarding the microbial world. The commensal organisms are, in fact, our allies in reversing the resistance problem. As they rebuild their constituencies, they will control the levels of resistance by out-competing resistant strains. Then, when bacteria or other microbes cause infections, they will be drug susceptible, and we will have the armamentarium to treat them effectively and successfully.
_
‘‘War is too important to be left to the generals,’’ Clemenceau famously said. In this context, war against microbes is too important to be left to the clinicians. This is not meant to be offensive; there are too many aspects of infection management that are entirely out of the scope and training of the clinician. Epidemiologists, pharmacologists (and, most importantly, experts in pharmacoeconomics), microbiologists, evolutionary, population, and molecular biologists all must participate in a concerted effort to design sound strategies to handle infectious diseases, including the use of antibiotics. Unfortunately, in developing countries, we do have only a few qualified experts in each field; and, on the other hand, the political control of most of these issues relies only upon physicians (or bureaucrats with medical training) who tend to think of all other disciplines as lesser fields. In any case, nonclinical experts need to get involved in this area, despite their natural aversion to clinical things; and clinicians need to allow others to join the fight against resistance. Building bridges is the first task. Antibiotic resistance is the consequence of a variety of biological, pharmacological, and societal variables that occur worldwide but that present themselves in the worst possible combinations in developing countries. Then we need to consider that infectious diseases are much more common here, as poor sanitary and work-safety conditions, starvation and malnutrition, lack of medical services (and an excess of ‘‘alternative medicine’’ options), and larger exposure to environmental agents that increase the likelihood of infection (e.g., weather changes, arthropod vectors) affect much more and a much larger fraction of the population than in developed countries. Conditions are only likely to get worse, as the divide between rich and poor countries widens, as it does between rich and poor people within poor countries, and also as climate change, war, and migration introduce entirely new variables to systems that were in an equilibrium of sorts for many years. To revert or at least stabilize things as they are now, we must start addressing the problem that affects particularly the poor countries: poverty itself. All other approaches are palliative at best.
_
Antimicrobial Resistance should not overshadow broader issue of Access to Medicines:
Any options in support of access to and stewardship of antimicrobials should balance monitoring, control and conservation on the one hand, and access and affordability on the other. From the development perspective, it would seem far too drastic to advocate for stringent measures of control such as limiting consumer access to antimicrobials, or withholding life-saving antibiotics for human use under the concept of conservation. It is necessary to avoid unnecessarily restrictive policies in particular in developing countries where the lack of access to antibiotics kills more than the resistance itself. Undue restriction on prescription and use of antibiotics, or the imposition of a ban on sales would result in greater barriers to access and could in practice undermine flexibilities provided for public health objectives in international agreements.
__
Evolutionary perspectives:
One way the evolution of antibiotic resistance can be slowed is by minimizing the strength of natural selection for resistance genes. This means reducing antibiotic use. Don’t treat asymptomatic infections. Use antibiotics only where they can work. 70% of acute bronchitis cases are treated with antibiotics to no effect. When treatment is necessary, use as little as possible. Decrease the need for antibiotic treatment with vaccines, hygiene, and isolation of infected patients. Prevent non-medical uses like growth promotion in farm animals. Reducing antibiotic use also limits selection for resistance in harmless bacteria that can donate genes to pathogens by HGT, as does targeting pathogens with narrow spectrum antibiotics. Another way to slow resistance evolution is to prevent pathogens acquiring resistance genes in the first place. In some cases, this can be done with high doses (dead bacteria can’t evolve) or combination therapy (acquiring resistance to several drugs at once is unlikely). However aggressive treatment also maximizes the evolutionary advantage of already resistant pathogens or of non-target bacteria that can be a source of resistance genes. Consequently, treatments designed to minimize the rate of resistance acquisition might not be best when resistance is already present. Bottom line is antibiotics should be used only when necessary and then, appropriately.
Other major research questions:
(i) when do drug cocktails select for multidrug resistance faster than sequential monotherapies?
(ii) does repeated antibiotic use generate a microbiomic reservoir of resistance genes for future pathogens?
(iii) what properties of particular drugs make them more evolution-proof than others?
(iv) is inappropriate antibiotic use the main evolutionary force undermining drug efficacy—or is it medically appropriate use?
________
Spatiotemporal microbial evolution on antibiotic landscapes: September 2016 study:
Researchers have used a meter-long petri dish to show the frightening speed at which bacteria can evolve and develop resistance to modern antibiotics. To better understand the emergence of antibiotic resistance in space and time, Michael Baym and colleagues developed a device called the microbial evolution growth arena plate, or MEGA plate — a large, rectangular petri dish across which different concentrations of antibiotics can be applied; here, the researchers used trimethoprim and ciprofloxacin. Bacteria were cultured at one location on the plate; as competition for resources increased, they spread to other regions. Applying varying levels of antibiotics, the authors were able to map out mutations that allowed increasingly resistant mutant bacteria to spread. They found it took bacteria just 11 days to spread from a section of the petri dish with very little antibiotic to a section with 1000 times the amount. The experiment shows just how easy it is for bacteria to evolve resistance. They divided petri dish into nine panels. The two outer panels had zero antibiotics. The two panels directly inside had antibiotics at a level where “E. coli can barely survive”. Every 13 centimetres inwards, they increased the amount of antibiotics. New mutations arise at each barrier where the concentration of antibiotic increases 10-fold. The evolutionary path through which this resistance arose was then mapped to understand how antibiotic resistance develops. The experiment shows just how easy it is for bacteria to evolve resistance – how quickly evolution can occur. In just 11 days, resistance levels increased by over 1000-fold. This shows that resistance does not evolve through any one big step but though a series of mutations, each one contributing just a bit and together accumulating to a very large increase in resistance. The researchers tracked which bacteria first developed antibiotic resistance at each level and then sequenced the genome of those bacteria to determine exactly what had changed. What’s more, when they set up a different MEGA plate, which went only between zero and a high concentration of antibiotic, the bacteria never developed antibiotic resistance. This showed the “moderately challenging” pressure of intermediate concentrations was actually essential to allow for resistance to develop. The most resistant bacteria were not even found on the front lines. Professor Kishony and his team sequenced the genomes of the bacteria for each new mutant and found that occasionally the bacteria behind the front line had developed extra, complementary mutations that made them better at surviving and thriving in the high-antibiotic environments. However, they couldn’t get to the tank’s frontlines because of all the other bacteria in the way. This demonstrated that it’s not just a matter of which bacteria can mutate the fastest, but which can do so in the right time and place to succeed. “We hope that by systematically mapping the mutations leading to resistance we will be able in the future to develop ‘anticipatory diagnostics’, which will tell the doctor which antibiotics to use and which combinations to use to best limit the chance of evolution,” said Professor Kishony. “Our approach to emerging antibiotic resistance definitely can’t just be to use incrementally more and more antibiotics,” he said. “We need to take antibiotics in the correct regimen and dose. Too-low levels generate these concentration steps that speed up evolution,” he added. “In general, we should be mindful about not using antibiotics unless really needed.” Mutations that increased resistance often came at the cost of reduced growth, which was subsequently restored by additional compensatory mutations, the authors found. Intriguingly, the spatial location of bacterial species played a role in their success in developing resistance. For example, when the researchers moved the trapped mutants (those behind their “fit” parents) to the “frontlines” of the culture, they were able to grow into new regions where the frontline bacteria could not. In light of this finding, the authors suggest that the fitness of bacterial populations is not driven by the fittest mutants, but rather by those that are both sufficiently fit and arise sufficiently close to the advancing front. The fittest mutants were not those most likely to infiltrate higher antibiotic concentrations. Instead, bacteria “behind” the very fittest on the growth plate became capable of surviving at highest antibiotic concentrations. The results provide important insights into the evolutionary patterns and mechanisms that drive bacteria’s success in overcoming antibiotics, a phenomenon that threatens human health worldwide. The value of this work is not a clinical model. In order to grow bacteria on a petri dish of that size, bacteria should be able to swim, which is something E. coli can do but many other model organisms cannot. In fact, the researchers themselves do not claim their MEGA plate is a mirror of how bacteria thrive in the world. Still this experiment offers a more closely related view of real environments than any previous studies, which relied on flasks and beakers filled with bacteria or even more sophisticated, though very small scale, microfluidic devices.
_______
The Race to reverse Antibiotic Resistance: Alternating Therapy: a 2016 study:
Alternating between different antibiotics could help steer bacterial evolution away from antibiotic resistance:
When a patient with a bacterial infection takes an antibiotic, the drug kills most of the bacteria. Some bacteria, however, may have genetic mutations that make them resistant to the drug’s effects. After all, bacteria reproduce quickly, which gives them plenty of chances to pick up a random mutation that turn out to be useful. Bacteria with the resistant mutation won’t die; instead, they’ll survive and produce more resistant bacteria. Soon, the antibiotic won’t be able to treat the patient’s infection. That process has been going on since the early 20th century, when the first commercial antibiotics hit the shelves. Doctors strike at bacteria with a drug, and bacteria evolve ways to resist. We need a new strategy, and some researchers say that means fighting evolution with evolution. By applying selective pressure that wipes out resistant strains instead of non-resistant ones, they say it may be possible to reverse the course of bacterial evolution by, in effect, selectively breeding bacteria into slightly more compliant enemies. These approaches can also be used in a ‘one-two punch’ treatment strategy. First select against resistance to eliminate single-drug resistant mutants from a population, and then follow up with the now-effective classical antibiotic.
_
For a while, some researchers thought that simply removing the selective pressure that produced resistant strains would be enough to reverse the trend. The idea is appealingly simple: The mechanisms that allow bacteria to resist antibiotics also cost the bacteria a lot of energy. Without the selective pressure that helped resistance evolve in the first place, bacteria that don’t put energy into unnecessary resistance mechanisms will grow and reproduce more quickly than those that do. However, it turns out that resistance isn’t as physiologically costly to maintain as some researchers once thought, so removing the pressure isn’t enough to cause drug-resistant bacteria to simply lay down their weapons. People have tried but it never works in any kind of reliable way. Instead, doctors are looking for a way to apply pressure that will actively select against resistant strains and in favor of non-resistant ones.
_
A genetic mutation that helps bacteria resist one antibiotic might also change something that leaves the bacteria vulnerable to another antibiotic. By adapting to the presence of one antibiotic, bacteria effectively specialize and can become less resilient to other antibiotics. If a pathogen is resistant to one drug, doctors can switch to another drug – one to which the pathogen is more vulnerable. When the new drug kills off the resistant bacteria, nonresistant versions will have a better chance of surviving. Alternating between antibiotics could lead to the evolution of strains that have lost their resistance to both drugs. That’s a simplified explanation, of course. In practice, drug cycling may involve several drugs, rather than just two. And that strategy depends on which mutations show up in the bacteria, and how effective they are at creating resistance. However, drug cycling can help steer bacterial evolution away from some types of resistance, if doctors choose the right drugs.
_
In a study published in 2015, Barlow, along with a team of mathematicians, tested 15 beta-lactam antibiotics against bacteria with different genotypes of an enzyme called beta-lactamase, which helps some bacteria resist beta-lactam antibiotics like penicillin. Based on the bacteria’s growth rates, the researchers measured how resistant each strain was to each antibiotic. Then, they predicted the probability of each antibiotic selecting for mutations that would decrease resistance. Based on that, they developed an algorithm to predict which sequence of five antibiotics would most likely reverse the bacteria’s evolution of resistance. One sequence of drugs had a 38 percent to 100 percent probability of coaxing the bacteria to evolve back to a non-resistant wild type, while another had a probability between 60 percent and 100 percent. Now, Barlow and her colleagues are studying how different concentrations of the drugs might affect those probabilities. They’re using miniature, synthetic organs and circulatory systems called “bodies-on-chips” to simulate how drugs would circulate in a person. According to Barlow, the simulations are a necessary step on the way to animal trials and, eventually, human clinical trials of the drug sequences recommended by her team’s algorithms. “It’s going to be important for patients in maybe five years,” she said.
________
Resistance, fitness and virulence of microorganism:
The selective pressure of antibiotics in general results in the emergence of resistance mechanisms on the part of bacteria. Hence, a direct correlation often exists between antibiotic use and the development of bacterial resistance to antibiotics over time. Conversely, the incidence of antibiotic resistance can be reduced by the restriction of antibiotic usage. This interrelation between antibiotic usage and resistance development is partly determined by the so-called biological cost of antibiotic resistance. Since antibiotic resistance often is characterized by detriments in biological fitness, resistant bacteria can be decimated selectively by a decreased use of antibiotics. It appears, however, that resistance never completely disappears. This feature may be owing to the fact that the loss of biological (Darwinian) fitness on the part of resistant bacteria can easily be overcome by the acquisition of compensatory mutations, thereby stabilizing the resistant bacteria within a given population. There is the price associated with antibiotic resistance from the perspective of the bacterium itself, to the extent that resistance determinants may interfere with the normal physiological process in the cell and hence cause a reduction in the level of biological fitness. In point of fact, several recent studies have shown that isolates with resistance genotypes are less fit than their sensitive counterparts in the absence of antibiotic selective pressure, thereby indicating a considerable cost of resistance. The biological cost of antibiotic resistance, however, can be considerably diminished and even compensated for by evolutionary changes within the bacterial genome. Hence, bacteria often do pay a metabolic price, such as reduced growth rate, reduced invasiveness, or loss of virulence, for the acquisition of drug resistance in the short term; but their adaptation to the physiological cost is likely to foster the stable maintenance of resistance in the long term. So antibiotic-resistant strains might also be fitter and more virulent, which may have profound impacts on the control and treatment of bacterial infections. Recent findings revealed a complicated love story between antibiotic resistance and bacterial virulence. There was an ancient paradigm about the ‘fitness cost of antibiotic resistance,’ but the emergence of the new technologies of high-throughput sequencing has changed the field, allowing researchers to study bacterial pathogenesis at the genome scale. This new, unbiased approached has revealed that unfortunately the worst case scenario of antibiotic resistant bacteria being more fit and virulent was not uncommon, particularly during infection. The situation complicates our fight against antibiotic resistance.
_
Increases in virulence often accompany acquisition of resistance: a 2002 study:
Helms et al. (2002) found that patients infected with pansusceptible Salmonella Typhimurium were 2.3 times more likely to die within 2 years after infection than persons in the general Danish population, and that patients infected with strains resistant to ampicillin, chloramphenicol, streptomycin, sulfonamide and tetracycline were 4.8 times (95% CI 2.2 to 10.2) more likely to die within 2 years. Furthermore, they established that quinolone resistance in this organism was associated with a mortality rate 10.3 times higher than the general population. Evidence is also mounting that, for some pathogens, increases in virulence often accompany acquisition of resistance. On the other hand, antibiotic resistant strains appear to be more virulent because we cannot kill them or stop their growth.
_
Biological Cost of Rifampin Resistance from the perspective of Staphylococcus aureus: a 2002 study:
Antibiotic resistance due to chromosomal mutations that cause structural modifications in the cellular target of the drug, such as rifampin resistance in Staphylococcus aureus, can be associated with a fitness burden. This study aimed to investigate the intrinsic detriments in biological fitness associated with RNA polymerase (rpoB) mutations that confer rifampin resistance in S. aureus. Three principal findings emerged. First, the competition assays of in vitro-selected Rifr mutants with their Rifs isogenic counterpart revealed that only one rpoB genotype displayed no fitness burden, whereas the other mutations were associated in some cases with a considerable loss of fitness. It must be kept in mind, however, that growth defects in vitro do not always necessarily go along with growth defects in vivo. Nonetheless, Moorman and Mandell also showed that rifampin-resistant strains of S. aureus display growth defects in vitro and in some cases display reduced virulence in an animal model. Second, no relationship between the magnitude of the biological cost and the level of resistance to rifampin could be detected. Third, the variation in frequency of rpoB mutations conferring resistance to rifampin in S. aureus in vivo appears to be a function of the Darwinian fitness of the organism. Indeed, the 481His→Asn mutation, which was not associated with a loss of fitness in vitro, was shown to be prevalent in 91% of the Rifr clinical S. aureus isolates tested. Moreover, since the in vivo isolates that display the 481His→Asn mutation were obtained from six countries and revealed different genetic backgrounds by macrorestriction analysis, it seems very unlikely that the high prevalence of this “no-cost” mutation can be explained by the clonal spread of a rifampin-resistant ancestral strain. As such, it seems likely that, in vivo, a functional restriction on RNA polymerase and subsequent bacterial fitness limit the expression of the full range of theoretical mutants defined in vitro. Interestingly, high-level resistance within clinical isolates was mainly attributable to double mutations within rpoB, including the mutational change 481His→Asn. This in turn is indicative of a resistance-mediating function on the part of the additional mutation, since the 481His→Asn mutational change on its own confers only low-level resistance. Regarding the two isolates that exhibited three mutational changes within rpoB in vivo, however, it still remains a matter of speculation as to whether both of the additional mutations, besides the 481His→Asn mutation, contribute to resistance. These findings demonstrate that the ability of resistance determinants to survive in bacteria is due not only to compensatory mutations that substantially diminish the biological cost of antibiotic resistance, as shown by others, but also to the selection among resistance alleles favoring those that impose the lowest (if any) biological costs, as proposed here. Another example of a no-cost resistance mutation has been elucidated in vitro for the rpsL gene, which is responsible for resistance to high concentrations of streptomycin in Salmonella enterica subsp. enterica serovar Typhimurium. Moreover, rpsL mutations responsible for streptomycin resistance in clinical isolates of Mycobacterium tuberculosis were shown to be the same as those with no cost in experiments performed with Salmonella serovar Typhimurium. Extrapolation of these findings makes it tempting to speculate that resistance will never disappear completely because there is no evolutionary disadvantage to being resistant once adaptation has taken place, i.e., by acquisition of compensatory mutations, or when fitness has not been diminished, i.e., by selection of no-cost mutations. Consequently, policies of decreased antibiotic usage intended to reduce resistance might not be as successful as originally anticipated. Finally, strict hygienic and antibiotic treatment regimens for prevention of the spread of resistance together with continued drug development are required if the human population wishes to stay one step ahead of antibiotic resistance.
_
A Fitness Cost associated with the Antibiotic Resistance Enzyme SME-1 β-Lactamase: a 2007 study:
The blaTEM-1 β-lactamase gene has become widespread due to the selective pressure of β-lactam use and its stable maintenance on transferable DNA elements. In contrast, blaSME-1 is rarely isolated and is confined to the chromosome of carbapenem-resistant Serratia marcescens strains. Dissemination of blaSME-1 via transfer to a mobile DNA element could hinder the use of carbapenems. In this study, blaSME-1 was determined to impart a fitness cost upon Escherichia coli in multiple genetic contexts and assays. Genetic screens and designed SME-1 mutants were utilized to identify the source of this fitness cost. These experiments established that the SME-1 protein was required for the fitness cost but also that the enzyme activity of SME-1 was not associated with the fitness cost. The genetic screens suggested that the SME-1 signal sequence was involved in the fitness cost. Consistent with these findings, exchange of the SME-1 signal sequence for the TEM-1 signal sequence alleviated the fitness cost while replacing the TEM-1 signal sequence with the SME-1 signal sequence imparted a fitness cost to TEM-1 β-lactamase. Taken together, these results suggest that fitness costs associated with some β-lactamases may limit their dissemination.
_
Association between antimicrobial resistance and virulence in Escherichia coli: a 2012 study:
The topic on the link on resistance/virulence is complex, considering the diversity of antimicrobial resistance genes, virulence factors, bacterial species and hosts. Most reports on this topic correlate the epidemiology of specific resistance genes with virulence genetic traits, a first step toward understanding whether there is a connection between resistance and virulence. More in depth molecular studies on the genetic between antimicrobial resistance and virulence determinants are sorely needed, to fully understand the interplay of resistance and virulence genes, whether virulence expression is affected by chromosomal mutations leading to specific resistance (e.g., fluoroquinolone resistance), if both determinants are inserted in the same mobile genetic element, like a conjugative plasmid, or the role of the phylogenetic background of the strain. Escherichia coli represents a major cause of morbidity and mortality worldwide. The treatment of E. coli infections is now threatened by the emergence of antimicrobial resistance. The dissemination of resistance is associated with genetic mobile elements, such as plasmids, that may also carry virulence determinants. A proficient pathogen should be virulent, resistant to antibiotics, and epidemic. However, the interplay between resistance and virulence is poorly understood. Despite the numerous controversies on this topic, findings from research published to date indicate that there is a link between resistance and virulence, as illustrated by the successful E. coli ST131 epidemic clone. Perhaps the most commonly accepted view is that resistance to quinolones is linked to a loss of virulence factors. However, the low virulent phylogenetic groups might be more prone to acquire resistance to quinolones. Specific characteristics of the E. coli genome that have yet to be identified may contribute to such genetic linkages. Research based on bacterial populations is sorely needed to help understand the molecular mechanisms underlying the association between resistance and virulence, that, in turn, may help manage the future disseminations of infectious diseases in their entirety.
__
Resistance and Fitness vis-à-vis malaria: a 2015 study:
One of the challenges to studying the interplay between parasite drug resistance and fitness is the lack of a direct measure for fitness. Comparison of relative growth rates in vitro or ex vivo is the commonly used approach, although growth rates may not represent relative fitness in the natural host. Assessment of parasite survival in the field provides an improved measure, although analyses are challenging. It has long been observed that P. falciparum genetic mutations that confer drug resistance are associated with altered biological fitness of the parasite. However, various investigators have reported both increased and decreased fitness in resistant parasites. Data from Uganda showed significantly lower prevalence of symptoms among children infected with parasites containing chloroquine resistance mutations compared with those infected with wild-type parasites, consistent with greater virulence for wild-type parasites.
_______
Emergence, Spread, and Environmental Effect of Antimicrobial Resistance: How use of an Antimicrobial Anywhere can increase resistance to any Antimicrobial Anywhere else: a 2002 study:
Use of an antimicrobial agent selects for overgrowth of a bacterial strain that has a gene expressing resistance to the agent. It also selects for the assembly and evolution of complex genetic vectors encoding, expressing, linking, and spreading that and other resistance genes. Once evolved, a competitive construct of such genetic elements may spread widely through the world’s bacterial populations. A bacterial isolate at any place may thus be resistant—not only because nearby use of antimicrobials had amplified such a genetic construct locally, but also because distant use had caused the construct or its components to evolve in the first place and spread there. The levels of resistance at any time and place may therefore reflect in part the total number of bacteria in the world exposed to antimicrobials up until then. Tracing the evolution and spread of such genetic elements through bacterial populations far from one another, such as those of animals and humans, can be facilitated by newer genetic methods. Estimates of the costs to human health of antimicrobial use in animals are often based on the direct infection of humans through contact with animals or animal food products by an epizootic pathogen such as Salmonella or Campylobacter. There is growing evidence, however, that antimicrobial-resistance genes and their genetic vectors, once evolved in bacteria of any kind anywhere, can spread indirectly through the world’s interconnecting commensal, environmental, and pathogenic bacterial populations to other kinds of bacteria anywhere else. The evidence for widespread indirect dissemination of antimicrobial resistance comes from varied sources, such as microbial population biology, microbial genetics, and clinical and epidemiological observations.
_
More than a billion trillion bacteria of diverse types live and compete on the world’s people and animals and in the environment. Hypothetically, any one of them might replicate every half hour to generate a billion progeny overnight; however, in reality, any given bacterium has only a 50–50 chance of replicating successfully. Each germ competes in a niche somewhere. A mutation in one of its thousands of enzymes might enable it to better use or tolerate something in the complex environment of that niche. By thus outgrowing its competitors even slightly, it could greatly amplify its progeny. Atwood et al noticed 50 years ago that mutants in a continuous culture of Escherichia coli would persist for hundreds of generations but abruptly disappear when a new advantageous mutation arose. They called this recurrent purging of diversity “periodic selection.” Periodic selection is limited, however, by niche diversity. A new strain with even a small advantage in one of many available ecological niches might sweep through that niche, as in the flask, but lack that same advantage in the different conditions of the next niche. Niche diversity thus tends to limit periodic selection in the real world. A strain of antimicrobial-resistant bacteria is a special case, however, because antimicrobial exposure affects all niches. In the presence of the agent, the strain has not a slight local advantage but rather a near-universal overwhelming advantage. Its competitors in all niches die, and its advantage overrides lesser niche-to-niche differences, enabling the strain to disseminate through many or all of the niches exposed to the agent. By thus diminishing effective niche compartmentalization, an antimicrobial agent has the potential to make a treated human, animal, or portion of the environment (or a group, as in an intensive care unit or feedlot)—or all antimicrobial treated hosts everywhere—more like a single flask.
______
Can we ever overcome AMR?
The global spread of microbial resistance is a predominant reason why infectious diseases have not been conquered. It is commonly expressed that physician misuse of antibiotics is the cause of antibiotic resistance in microbes and that, if we could only convince physicians to use antibiotics responsibly, we could “win the war against microbes.” Unfortunately, this belief is a fallacy that reflects an alarming lack of respect for the incredible power of microbes. We are weak in comparison with the adaptability of microbes, which inhabit literally every possible climate and environment on the planet, despite extremes of boiling or freezing temperatures, pressures sufficient to crush virtually any human-made submersible, extreme salinity, zero oxygen content, presence or absence of sunlight, etc. Indeed, from the microbial perspective, human beings are nothing more than walking microbial planets; there are 5–10 times more microbes living on and in every human being than there are human cells in our bodies. Bacteria even exist in large numbers miles deep in the midst of solid rock in the earth’s crust. Because of this extraordinary diversity of habitat, microbes comprise fully 60% of the biomass on the planet (90% if cellulose is excluded from the calculation), despite their submicron size. Microbes have had 3.5 billion years to adapt to the various environments on planet Earth. The power that drives microbial adaptability is genetic plasticity and rapid replication. It takes many bacteria only 20–30 min to replicate; it takes human beings 20–30 years to replicate. Given the above, there is no doubt that microbes are the most numerous, diverse, and adaptable organisms that have ever lived on the planet. On reflection, perhaps it would be wise to reconsider the frequently used metaphor of humans being “at war with microbes”. It is absurd to believe that we could ever claim victory in a war against organisms that outnumber us by a factor of 1022, that outweigh us by a factor of 108, that have existed for 1000 times longer than our species, and that can undergo as many as 500,000 generations during 1 of our generations. Furthermore, the weapons in a war against microbes would be antibiotics. We need to remember that human beings did not invent antibiotics; we merely discovered them. Genetic analysis of microbial metabolic pathways indicates that microbes invented both β -lactam antibiotics and β -lactamase enzymes to resist those antibiotics >2 billion years ago. In contrast, antibiotics were not discovered by humans until the first half of the 20th century. Thus, microbes have had collective experience creating and defeating antibiotics for 20 million times longer than Homo sapiens have known that antibiotics existed.
_
From this framework, it is obvious that microbes do not need our help in creating antibiotic resistance. On the other hand, what human beings can do is affect the rate of spread of bacterial resistance by applying selective pressure via exposure to the thousands of metric tons of antibiotics we have used in patients and livestock over the past half century. Methods to control unnecessary use of antibiotics include appropriate regulations on use of antibiotics in agriculture (including elimination of use of antibiotics to promote growth of food animals), restriction of antibiotic use to pathogen-specific agents, and limits on the common practice of using antibacterial agents for viral infections. Clearly, it is desirable to use antibiotics only when appropriate, to try to limit selective pressure that increases the frequency of resistance. Nevertheless, the distinction between causality of microbial resistance and the rate of spread of resistance must be recognized if we are to create a true solution to the problem of antibiotic resistance. If our misuse of antibiotics causes drug resistance, the solution that would allow us to forever defeat microbial resistance would be for us to strictly use antibiotics only when truly indicated. On the other hand, if our misuse of antibiotics affects the rate of spread of resistance but does not actually cause resistance, then using antibiotics correctly will not stop microbial resistance, it will only slow it down so that we can find a real solution to the problem. Framed in this context, it is clear that convincing physicians to use antibiotics properly is an important step to take, not because it is a solution to drug resistance, but because it will buy us more time to create a real solution to the problem. Ultimately, we must concede that we will never truly defeat microbial resistance; we can only keep pace with it. The only viable, long-term solution to the problem of microbial resistance is to have in place in perpetuity a continuing, steady development of new antibiotics and other strategies (including immunotherapeutics and vaccines, diagnostics and antibiotic stewardship programs to improve targeted therapy, and well-coordinated and -funded domestic and international monitoring, tracking, and prevention and control plans) to respond to new drug-resistant threats. Finally, because it takes years to develop a new drug, planning must include consideration of needs that are immediate as well. These concepts have been summarized succinctly and precisely by Nobel Prize winner Dr. Joshua Lederberg, who stated, “The future of humanity and microbes will likely evolve as… episodes of our wits versus their genes”. I hope that our wits will overcome their genes.
__________
__________
Moral of the story:
1. Antimicrobial drugs include all drugs that work against a variety of microorganisms, such as bacteria, viruses, fungi, and parasites. An antibiotic drug is effective against bacteria. All antibiotics are antimicrobials, but not all antimicrobials are antibiotics. However, the terms ‘antibiotics’ and ‘antimicrobials’ are often used synonymously. Antibiotics work synergistically with immune system to kill microbes or inhibit growth of microbes.
_
2. Antimicrobial resistance (AMR) is the ability of a microorganism strain to survive and/or to multiply despite the administration and absorption of an antimicrobial given in doses equal or higher than those usually recommended, but within the limits of tolerance of the patients. This broad term also covers antibiotic resistance, which applies to bacteria and antibiotics. It is really only considered “resistance” when it occurs in a microorganism that used to be susceptible to an antimicrobial’s effects but now is not susceptible; this doesn’t really apply to a microorganism that was never susceptible to that antimicrobial. Antimicrobial resistance should be distinguished from other causes of drug failure such as non-adherence, insufficient drug levels, inability to reach sites of infection, presence of foreign body making successful antibiotic treatment more difficult and drug regimens with intrinsically weak antimicrobial activity.
_
3. Genetic analysis of microbial metabolic pathways indicates that microbes invented both β -lactam antibiotics and β -lactamase enzymes to resist those antibiotics about 2 billion years ago. Paleontological data show that both antibiotics and antibiotic resistance are ancient compounds and mechanisms. Human beings did not invent antibiotics; they merely discovered them.
_
4. Antibiotic tolerance is distinct from antibiotic resistance. Antibiotic tolerance is not caused by mutant microbes, but rather by microbial cells that exist in a transient, dormant, non-dividing state. These dormant cells survive the antimicrobial treatments that kill the majority of their genetically identical siblings. These dormant cells cannot be eliminated by the immune system and thereby cause relapsing and chronic infections. These dormant cells resume growth when the antimicrobial agent is removed, and their progeny is sensitive to antimicrobial agents.
_
5. Although antimicrobial resistance in general is posing grave problems for public health, resistance among bacteria poses far greater threat than other microbes because of various reasons: (a) the abuse of antibacterial drugs is much higher than that of antifungal or antiviral agents; the later ones are seldom self-prescribed, wrongfully used as prophylaxis, or have agricultural usage; (b) bacterial genetic characteristics and abilities enable a rapid evolution toward resistance in ways that exceed by far those of viruses, fungi, and protozoa: haploidy, horizontal gene transfer mechanisms, extrachromosomal elements are all features that foster resistance and that are almost unique to bacteria; (c) bacteria appear to be much more abundant than viruses, fungi, and protozoa as microbiota of humans, which increases exponentially the exposure of the former to antibiotics each time they are used clinically, creating more chances of resistance to emerge and be selected; (d) bacterial diseases are also more abundant, at least for treatment purposes, increasing also the exposure to antibacterial drugs, perhaps with the exception of malaria.
_
6. The abilities of microorganisms to utilize the various strategies to resist antimicrobial compounds are all genetically encoded. Bacteria may be intrinsically resistant to some class of antimicrobial agents, or may acquire resistance by de novo mutation or via the acquisition of resistance genes from other organisms. Intrinsic resistance is due to innately coded genes which create natural “insensitivity” to a particular antibiotic. Innate resistance is normally expressed by virtually all strains of that particular bacterial species. Acquired resistance is gained by previously susceptible bacteria either through mutation or horizontally obtained from other bacteria possessing such resistance via transformation, transduction or conjugation. Acquired resistance is limited to subpopulations of a particular bacterial species and may result from selective pressure exerted by antibiotic usage.
_
7. Spontaneous mutations in bacteria are random which may confer antibiotic resistance; they do not arise in response to exposure to the antibiotic. Antibiotic exert selection pressure so that in any large population of bacteria, few cells will be present which possess random mutations conferring resistant traits enabling them to survive in the presence of antibiotic, which then divide rapidly and produce a billion offspring in a single day thereby transfer their genetic material containing resistance genes to offspring. This spread of resistance is an outcome of natural selection and should be viewed as an expected phenomenon of the Darwinian biological principle of “survival of the fittest.”
_
8. Once bacteria acquires resistant gene by mutation, it is passed on to its progeny by vertical gene transfer. Once bacteria acquires resistant gene by horizontal gene transfer, it is passed on to other bacteria of same species and different species by horizontal gene transfer. Bacteria can also transfer resistant gene plasmids acquired through horizontal gene transfer vertically to its progeny by fixing plasmid gene in its genome. I hypothesize that resistance gene acquired through mutation which is transferred to progeny by vertical gene transfer; may replicate a copy that may be incorporated in the bacterial plasmid to be transmitted to other bacteria by horizontal gene transfer. In other words, at some point, vertical gene transfer and horizontal gene transfer merge with the sole objective of survival.
_
9. Commensal is a microorganism which typically does not cause disease in an individual but may cause disease when it travels out of its natural habitat in the same individual or travel to another individual or develop pathogenic strain. Excepting tuberculosis, majority of bacterial illness results from infection by commensal organisms, such as Escherichia coli, streptococci, and staphylococci. The two roles that commensal bacteria play vis-à-vis antibiotic resistance are as reservoirs of antibiotic resistance genes that can be transferred to pathogens via horizontal gene transfer and as antibiotic resistant opportunistic pathogens. The intestinal microbiota forms the main reservoir of multidrug-resistant gram negative bacterial infections in critically ill patients. The commensal organisms are, in fact, our allies in reversing the resistance problem. It is our duty to see that they do not develop resistance by inappropriate use of antibiotics; so that they build their constituencies to control the levels of resistance by out-competing resistant strains. The fastest way to eliminate resistant strains is to outnumber them with susceptible strains. Another way inappropriate antibiotic use can harm is by killing commensal organism and thereby reducing colonization resistance and promoting growth of new and often harmful microorganisms. Also, killing commensal bacteria with antibiotics might well weaken the immune system.
_
10. Remember bacteria are “promiscuous”. They commingle genes with their own kind or with fairly unfamiliar strangers, widely distributing resistance genes. Certain antibiotic resistance genes are easily transferred from one bacterial species to another, and can move between farm animals and the human gut which causes spread of antibiotic resistance. The long-term use of a single antibiotic (that is, for more than 10 days) will select for bacteria that are resistant not only to that antibiotic, but to several others. This phenomenon reflects the linkage of different resistance genes on the same transposon or plasmid. MDR plasmids eventually emerge with the prolonged use of a single antimicrobial agent. Bacteria that are already resistant to one antibiotic seem to be favored in recruiting additional resistance traits from other bacteria sharing the environment. The evidence suggests that antibiotic resistance genes in human bacterial pathogens originate from a multitude of bacterial sources, indicating that the genomes of all bacteria can be considered as a single global gene pool into which most, if not all, bacteria can dip for genes necessary for survival. We have already discovered existence of more than 20,000 potential resistance genes (r genes) of nearly 400 different types, predicted from available bacterial genome sequences. Use of an antibiotic anywhere can increase resistance to any antibiotic anywhere else.
_
11. Bacteria can evolve resistance quickly and easily depending on selection pressure exerted by antibiotics. Global consumption of antibiotics in human medicine rose by nearly 40% between 2000 and 2010. It is estimated that as much as 50 percent of all antibiotic use is inappropriate, unnecessary or suboptimal leading to increased antimicrobial resistance. Rising antibiotic resistance can be attributed to use of antibiotics: in the human population; in the animal population; in the agriculture and spread of resistant strains between human or non-human sources. The overuse of antibiotics clearly drives the evolution of resistance. Wide-spectrum antibiotics apply selective pressure, not only upon the etiological agent of the infectious episode but also upon a larger fraction of the patient’s microbiota. As far as antibiotic resistance is concerned, every individual is at the mercy of others. Antibiotics are different than other drugs. If someone were to take a statin, it’s not going to diminish statin’s effectiveness on others. But with antibiotics, you can get an infection by drug-resistant bacteria even if you’ve never misused antibiotics in your life. So you are paying price for misuse of antibiotics by others. Besides antibiotics, use of disinfectants, antiseptics and antibacterial soap also causes antimicrobial resistance.
_
12. Discerning whether an infection is viral or bacterial can be challenging. This uncertainty accounts for much of the overuse and over-prescribing of antibiotics. Procalcitonin level, lipocalin level and molecular diagnostic tests (genomic sequencing) can help differentiation of bacterial infections from viral infections. The complete blood count (CBC) alone does not have adequate sensitivity or specificity to differentiate bacterial from viral infections although white blood cell count more than 15,000 and/or absolute neutrophil count more than 10,000 per microliter is more likely to be bacterial infection.
_
13. Antibiotic resistance is the consequence of a variety of biological, pharmacological, and societal variables that occur worldwide but that present themselves in the worst possible combinations in developing countries. AMR prevalence is 12% in high-income countries, 31% in upper-middle income countries and 78% in lower-middle income countries.
_
14. Antimicrobial resistance poses a fundamental threat to human health, development, and security. With the growth of global trade and travel, resistant microorganisms can spread promptly to any part of the world.
_
15. About 70 percent of the bacteria that cause infections in hospitals are resistant to at least one of the drugs most commonly used for treatment. The death rate for patients with serious infections caused by common resistant bacteria treated in hospitals can be about twice than that of patients with infections caused by the same non-resistant bacteria.
_
16. Antimicrobial resistant microorganisms usually benefit from extra time to multiply resulting in complications and spread to other organs. AMR reduces the effectiveness of treatment; thus patients remain infectious for a longer time, increasing the risk of spreading resistant microorganisms to other people.
_
17. Over all, antimicrobial resistance is associated with higher mortality rate, longer hospital stay, delayed recuperation, long term disability, increases health care costs as well as the economic burden on families and societies. AMR claims about 700,000 lives a year globally but by 2050 AMR could be responsible for 10 million deaths per year and at least $100 trillion (more than the size of the current world economy) in lost output. And these figures are conservative. However, many studies that attempt to estimate the clinical and the economic impact of infections caused by resistant microorganisms have fundamental flaws in their methodology.
_
18. Health care facilities, particularly those which are large and care for the most complex patients are a focal point in the emergence of antimicrobial resistance. Hospital infection control, hospital antibiotic policy, hospital antibiotic stewardship, hospital antibiogram and surveillance of antibiotic-resistant infections are keys to prevent/reduce AMR in hospitals.
_
19. It is found that public has minimal awareness of antibiotic resistance, antibiotics and infections. They want quick symptomatic relief. About 75 % people are ignorant about antibiotic resistance and believe that a patient taking drug becomes resistant to their effects, not the microbes. This ignorance and misunderstanding leads to inappropriate use of antibiotics by people.
_
20. Animals serve as mediators, reservoirs and disseminators of resistant bacterial strains and/or AMR genes. More antibiotics are used in poultry, swine and cattle to promote growth and prevent disease than are used by the entire human population. Consequently there is an increasing prevalence of antibiotic-resistant bacteria in livestock, poultry and aquaculture, which is transferred to humans by consumption of animal products, contact with animals and environment. One cannot overlook the fact that antibiotics, namely tetracyclines and streptomycin, are provided as pesticides for agriculture to be sprayed on to fruit trees. The environment, soil and sewage are also reservoirs of antibiotic resistance genes among both pathogen and commensal bacteria which can be transferred to humans.
_
21. In tuberculosis resistance emerge in treated hosts through mutation; while in many other important pathogens resistance is acquired by horizontal transfer of gene, so there resistance in the treated host is a relatively rare event. Therefore for tuberculosis, prevention of antimicrobial resistance in individual hosts is a primary method of preventing the spread of resistant organisms in the community.
_
22. Poor medication adherence is the main cause of HIV drug resistance.
_
23. Regions with low transmission and low immunity have high prevalence of resistant malaria (e.g. Southeast Asia) as compared with high transmission and partial immunity regions (e.g. Africa). Low transmission and low immunity cause high parasitemia coupled with indiscriminate use of different antimalarial drugs led to resistant malaria.
_
24. Genotypic (molecular) testing for AMR is better than conventional antibiotic sensitivity tests because genotypic testing is generally rapid and sensitive resulting in early detection of AMR allowing clinicians to change drugs quickly before bacterial/viral load increases and patient becomes severely ill.
_
25. Local surveillance of resistant microorganisms has become matters of major importance for the selection of appropriate therapeutic schemes and the implementation of infection control measures. At a national level, data on resistance and antibiotic use inform policy decisions, such as antibiotic guideline development or revision, and identify priorities for public health action, such as education campaigns or regulatory measures. Data from various national and global surveillance studies indicate that the incidence of antimicrobial-resistant pathogens is increasing. Surveillance and reporting of AMR and antibiotic usage is central to prevention and containment of AMR.
_
26. Preventing infections from happening in the first place through better hygiene, access to clean water, better sanitation, vector control, infection control in health-care facilities, and vaccination reduce the need for antibiotics. Hand washing has emerged as one of the most important measure to prevent infection and consequently prevent AMR.
_
27. Antibiotics should be used only when necessary and then, appropriately in humans and animals. When both agents are active, the benefit of combination therapy over adequate monotherapy has not been proven in terms of clinical cure or microbiological eradication. Once susceptibility testing results are known, monotherapy with the most active antibiotics could be considered.
_
28. It is the “moderately challenging” pressure of intermediate concentrations of antibiotic that is essential to allow for resistance to develop and not high concentration. Higher dosage and shorter duration rapidly reduce the total population of bacteria and cut the use of antibiotics and reduce AMR as shorter the duration of treatment, lower the selection pressure for resistance is in the patient. However such aggressive treatment also maximizes the evolutionary advantage of already resistant pathogens or of non-target bacteria that can be a source of resistance genes. Consequently, high dose short duration treatment designed to minimize the rate of resistance acquisition might not be appropriate when resistance is already present. So high dose short duration aggressive antibiotic therapy should be given only if you know that bacteria causing infection are susceptible.
_
29. Alternating between antibiotics could lead to the evolution of strains that have lost their resistance to alternating antibiotics. Alternating therapy is a proposed method in which two or three antibiotics are taken in a rotation versus taking just one antibiotic such that bacteria resistant to one antibiotic are killed when the next antibiotic is taken. Studies have found that this method reduces the rate at which antibiotic resistant bacteria emerge in vitro relative to a single drug for the entire duration.
_
30. The development of new antibiotics by the pharmaceutical industry, a strategy that had been effective at combating resistant bacteria in the past, is essentially stalled. Antibiotic development is no longer considered to be an economically wise investment for the pharmaceutical industry. 20 years after new antibiotic research has begun, the pharmaceutical company can break-even. Break-even is the point of balance making neither profit nor loss. In 2015, only six of the world’s top 50 pharmaceutical companies are still pursuing antibiotic research. The consequences of the failure to create new antibiotics could be catastrophic. Availability of effective antibiotics has revolutionized public health and has been responsible for enabling countless advancement in medical care.
_
31. No matter how many new antibiotics come out, if we continue to misuse them they might as well have never been discovered because inappropriate and unnecessary use of antibiotics is key to generate antibiotic resistance. Preventing resistance to existing antibiotics should take priority over developing new drugs, since they will also fail if the causes of resistance are not tackled. Rational use of older antibiotics could represent an alternative to the treatment of MDR bacterial pathogens as development of new antibiotic is slow and scarce, and resistance to the new antibiotics is also emerging. Another approach to the antibiotic resistance problem could be to design drugs that self-destruct after treatment, thereby removing a contributing factor in the propagation of resistance.
_
32. Principles of antimicrobial stewardship and appropriate use should be incorporated into undergraduate and postgraduate medical education in all medical colleges. Social media platforms – including Facebook and Twitter provide an effective method to reinforce antimicrobial stewardship programs (ASP) and encourage the use of ASP resources to promote antimicrobial mindfulness among all medical students and doctors.
_
33. Banning the sale of antibiotics without medical prescription to control AMR could be counter-productive as many populations of developing world lack access to medical care and such a ban would be denying access to life-saving drugs. In developing countries lack of access to antibiotics kills more than the resistance itself. Also, antibiotics prescribed by doctors are inappropriate in 50 out of 100 prescriptions.
_
34. Antibiotic reduction campaigns do not necessarily reduce resistance because:
a) Although selective pressure of antibiotics in general results in the emergence of resistance mechanisms on the part of bacteria, the converse is not true. Loss of resistance is slow even in the absence of the selecting antibiotic. This phenomenon reflects the minimal survival cost to the emerging resistant strains. Resistance will never disappear completely because there is no evolutionary disadvantage to being resistant once adaptation has taken place, i.e., by acquisition of compensatory mutations, or when fitness has not been diminished, i.e., by selection of no-cost mutations. Also, resistance genes are often linked with genes specifying resistance to other antimicrobials or toxic substances on the same plasmids. The presence of MDR plasmids assures maintenance of the plasmid as long as any one of the resistances provides a survival advantage to the host bacterium. Consequently, policies of decreased antibiotic usage intended to reduce resistance might not be as successful as originally anticipated.
b) Success of antibiotic reduction programs depends on which antibiotics are reduced as some select more strongly for resistance than others.
_
35. Antibiotic resistance may impose a biological cost, thereby reducing fitness of resistant strains, which can limit the growth and spread of resistant bacteria. However, additional mutations may compensate for this fitness cost and can aid the survival of these bacteria. Bacteria often do pay a metabolic price, such as reduced growth rate, reduced invasiveness, or loss of virulence, for the acquisition of drug resistance in the short term; but their adaptation to the physiological cost is likely to foster the stable maintenance of resistance in the long term. The biological cost of antibiotic resistance can be considerably diminished and even compensated for by evolutionary changes within the bacterial genome. The ability of resistance determinants to survive in bacteria is due not only to compensatory mutations that substantially diminish the biological cost of antibiotic resistance, but also to the selection among resistance alleles favoring those that impose the lowest (if any) biological costs. So the worst case scenario of antibiotic resistant bacteria being more fit and virulent is not uncommon, particularly during infection. Evidence indicates that at least in certain strains of E.Coli, a correlation exists between virulence and antimicrobial resistance due to presence of resistance gene with virulence determinant on the same plasmid that disseminate resistance. On the other hand, data from malaria study showed significantly lower prevalence of symptoms among children infected with parasites containing chloroquine resistance mutations compared with those infected with wild-type parasites, consistent with greater virulence for wild-type parasites. Another study found that fitness costs associated with some β-lactamases may limit their dissemination.
_
36. For certain pathogens, resistance to a particular antimicrobial is never seen. For example, in spite of decades of extensive use of penicillin, group A streptococci have never developed resistance to penicillin barring very few strains. This observation that continuing susceptibility has occurred despite the development of resistance to other antimicrobial agents demands explanation. It could be because despite millions of spontaneous random mutations, none could code for biochemical pathway to resist penicillin. It is because its genome is built in such a way that even millions of mutation cannot code for such pathway. I would call it genome incompatibility to resistance for a specific antibiotic. Also, the same factor of genome incompatibly to resistance prevents acquisition of penicillin resistance through plasmids. In other words, there are genetic factors in bacteria which prevent acquisition of specific resistance gene through plasmid. In other words, acquisition of resistance genes either by mutation or by plasmid is genetically correlated. This is a hypothesis which needs to be confirmed or rejected by experiments. If we could find genome code that confers incompatibility to resistance, we can introduce it in other bacteria so that they become sensitive to penicillin. Genome incompatibility to resistance for a specific antibiotic (exclusive sensitivity) is exactly opposite of intrinsic resistance due to innately coded genes which create natural “insensitivity” to a specific antibiotic. Of course ‘exclusive sensitivity’ is against evolutionary instinct to survive and therefore a ‘mistake’ evolutionary biologically. Time will tell whether a ‘mistake’ is corrected by converting ‘exclusive sensitivity’ to insensitivity.
_
37. Antibiotic resistance genes have the potential to be used for bio-terror purposes through genetically modified organisms.
_
38. India has indeed become epicentre of AMR due to following reasons:
a) Antibiotic use is a major driver of resistance. India is the world’s largest consumer of antibiotics for human health.
b) Over-the-counter, non-prescription sale of carbapenems is highest in the world in India and contribute to growing carbapenem resistance among Gram-negative organisms. India is afflicted by easy access to the strongest of antibiotics without prescriptions or diagnoses; by qualified doctors, not just quacks, who prescribe drugs with little thought; and by hospitals where overuse, misuse and inappropriate doses have created colonies of superbugs.
c) Release of untreated waste products containing active antibiotic ingredients from many substandard Indian pharmaceutical factories contaminate air, water and terrains that acts as a driver for the development of drug resistance and create environmental ‘reservoirs’ of antibiotic-resistant bacteria. Once established in the environment, the resistant bacteria can exchange genetic material with nearby bacteria and then spread around the world through air and water, and by travellers.
d) Growing wealth where many people are demanding antibiotics for minor infections coupled with cheap antibiotics result in inappropriate use of antibiotics.
e) India lags behind on basic public health measures like immunization, clean drinking water, vector control and sanitation; coupled with overcrowding, malnutrition, poor personal hygiene and an excess of ‘alternative medicine’ options result in high rates of infectious diseases which necessitates greater antibiotic use. Also, antibiotics continue to be prescribed or sold for diarrheal diseases and upper respiratory infections for which they have limited value.
f) Incomplete course or dose due to poverty in rural area and self-medication in urban area are common.
g) Excessive and unregulated usage of antibiotics in livestock and animal food production contributes to resistance in microorganisms.
h) Most Indian hospitals have no antibiotic policy, no antibiotic stewardship and no infection control specialists.
______
Dr Rajiv Desai. MD.
November 20, 2016
______
Postscript:
Every truth passes through three stages before it is recognised. In the first it is ridiculed, in the second it is opposed, in the third it is regarded as self-evident. It is time for India to accept the truth that it has indeed become epicentre of antimicrobial resistance. AMR is far more serious threat than conditions such as Zika and Ebola, which receive extensive media coverage.
______


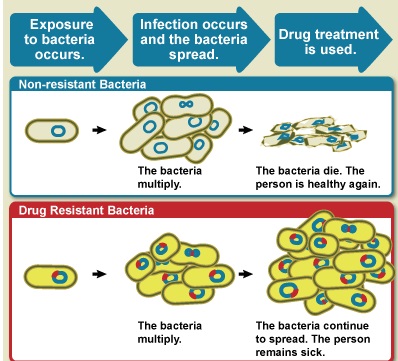
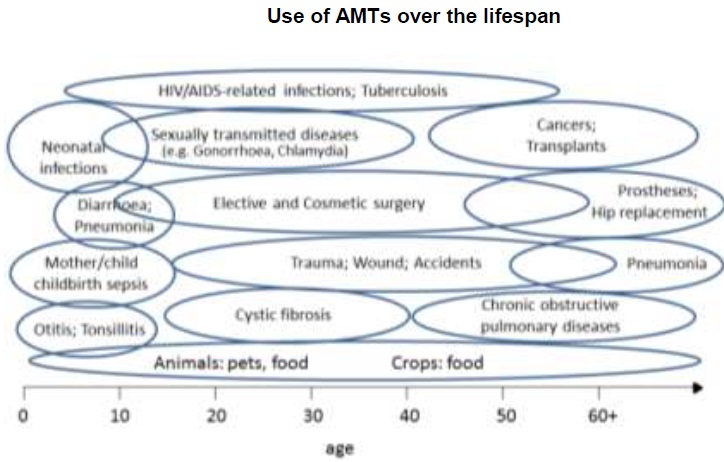
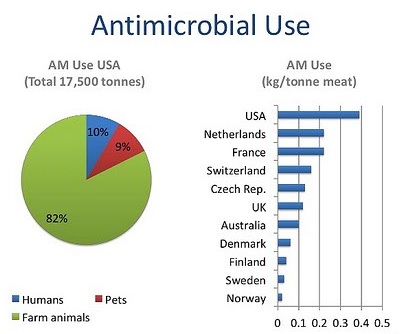
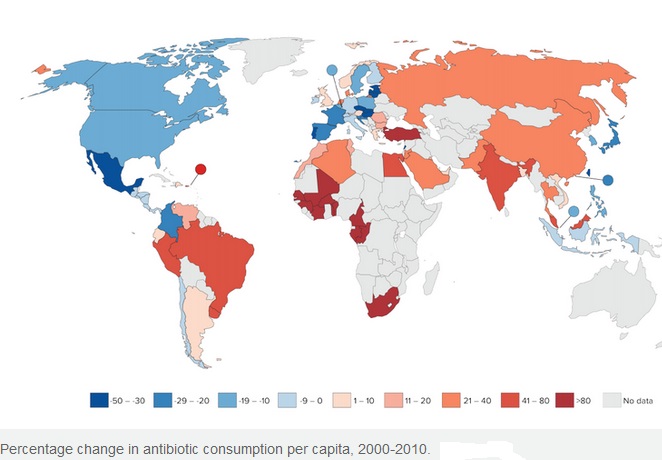
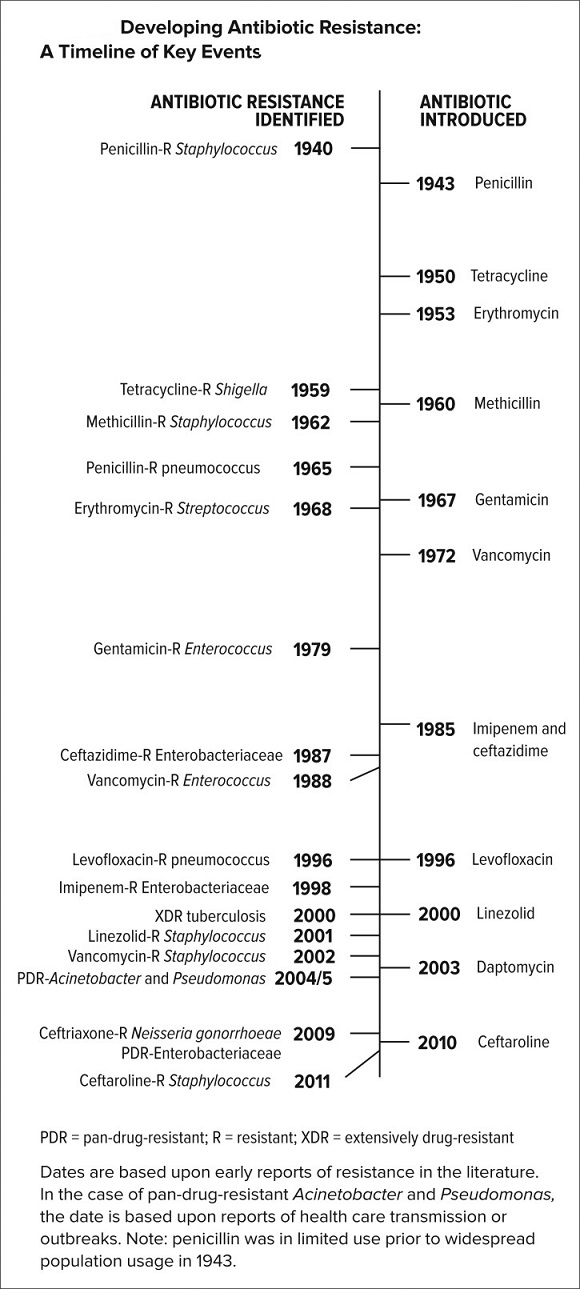

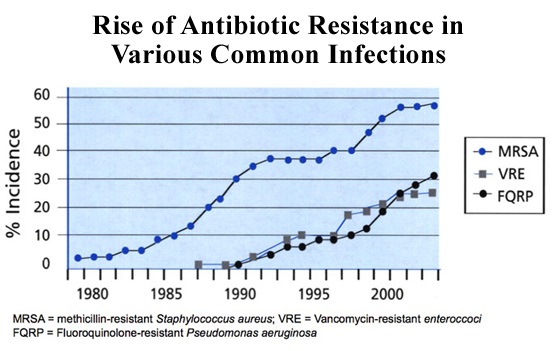
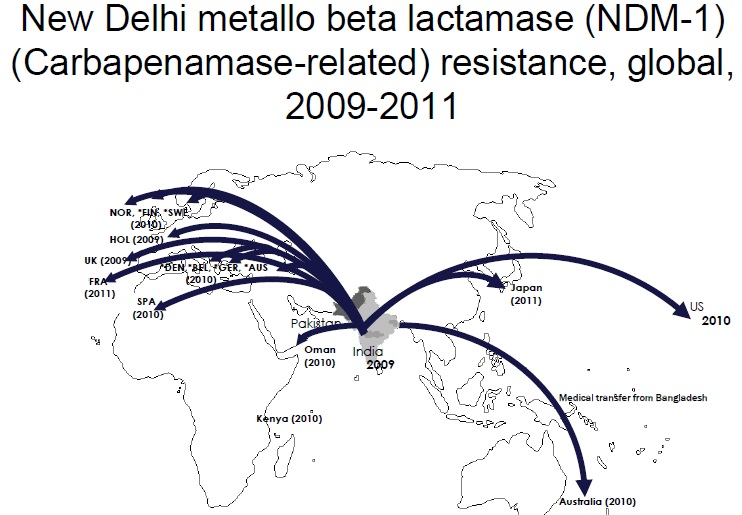
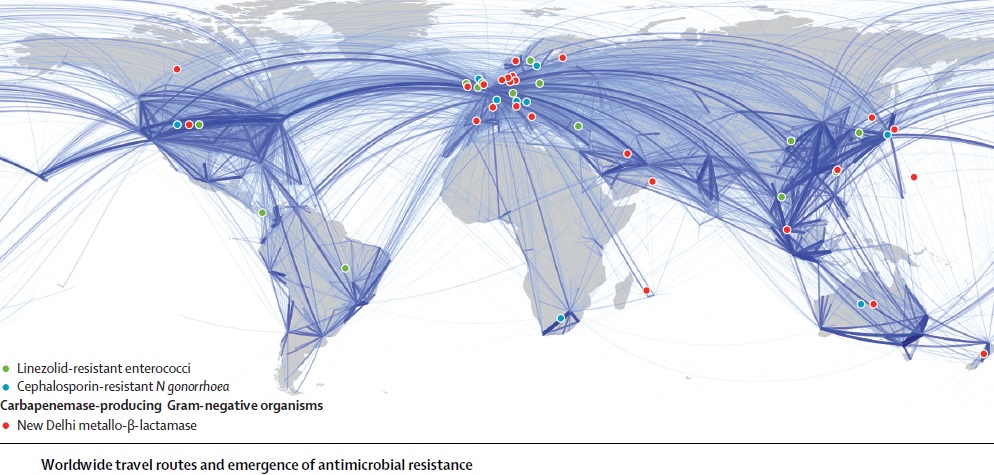

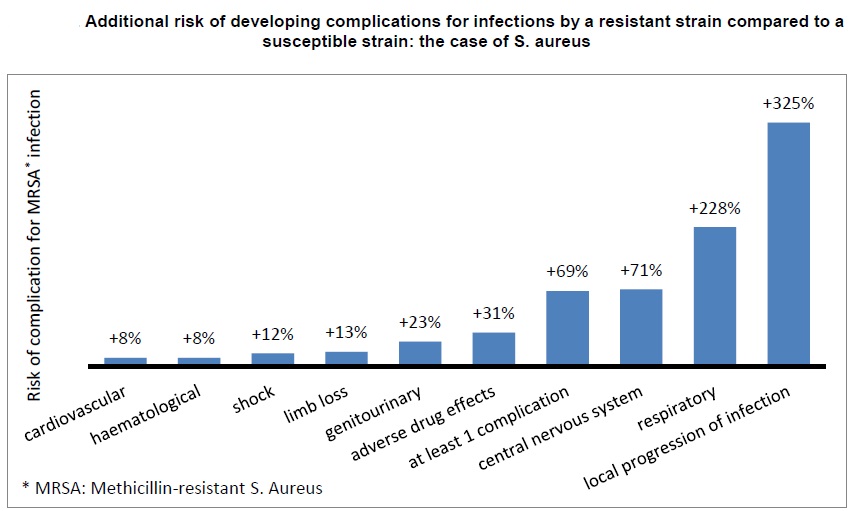

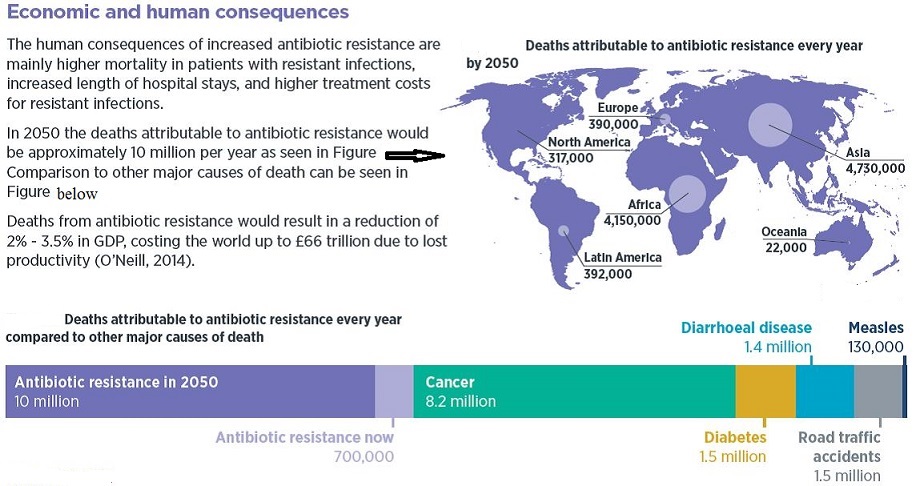
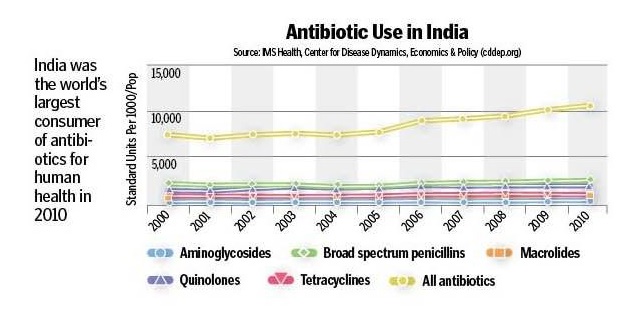
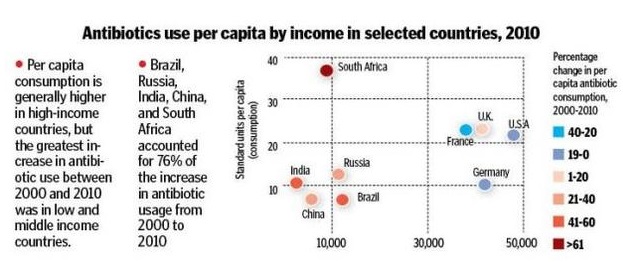
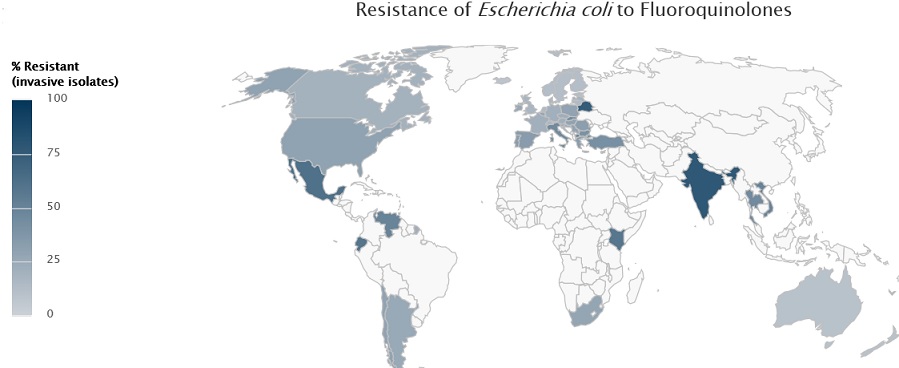
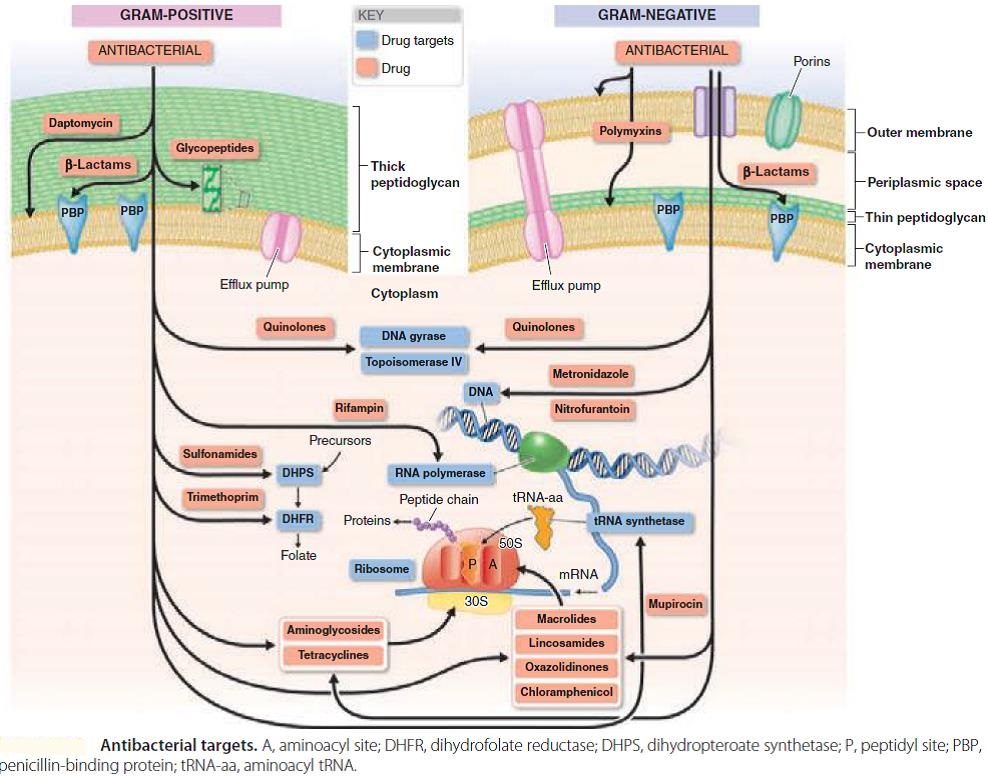
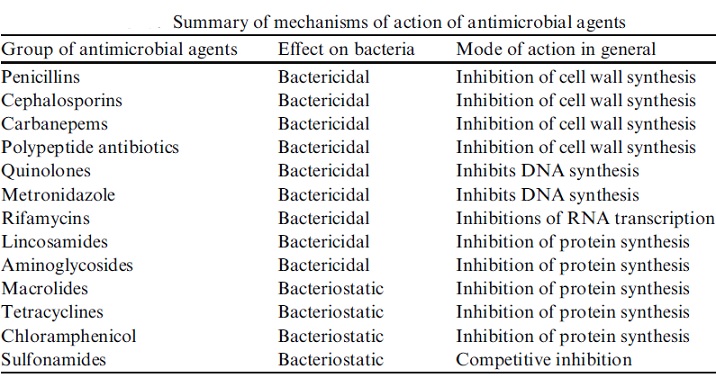
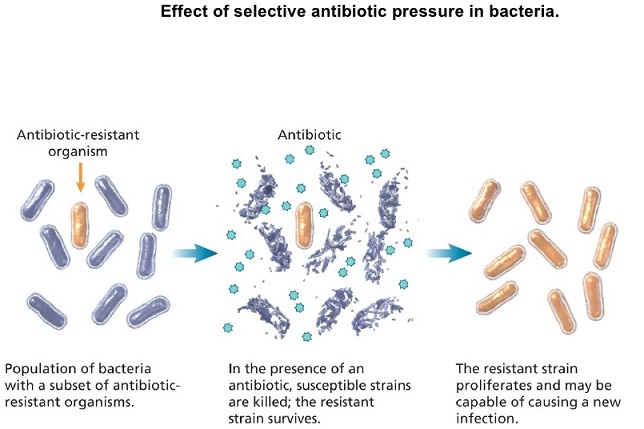
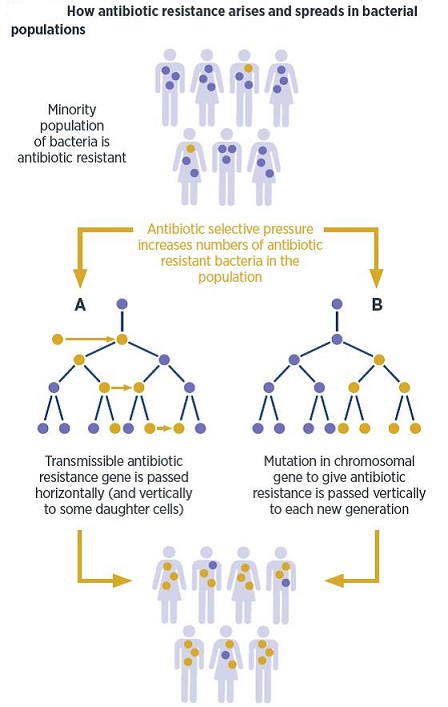
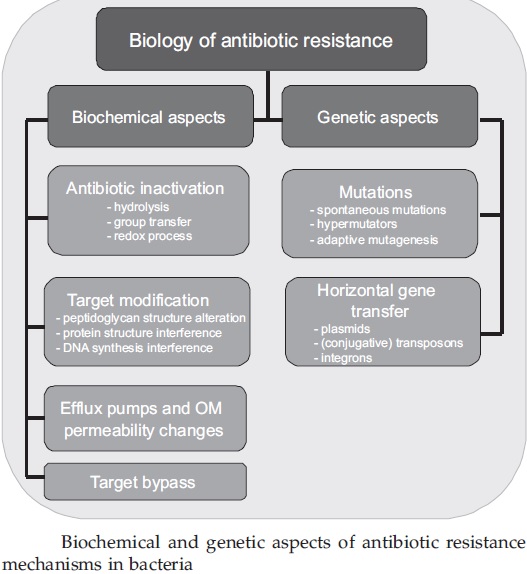
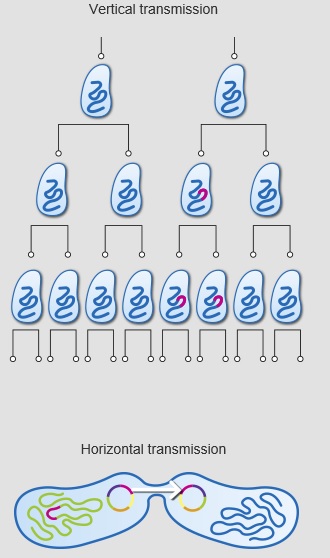
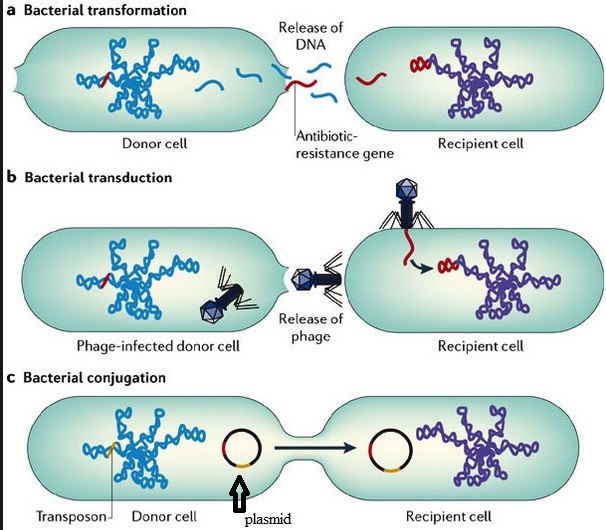
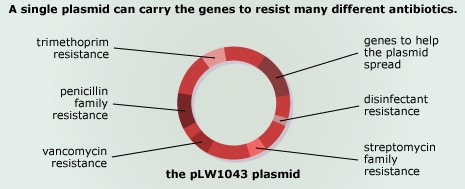
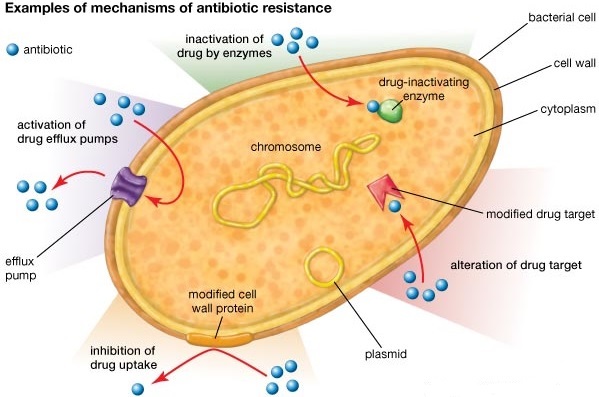
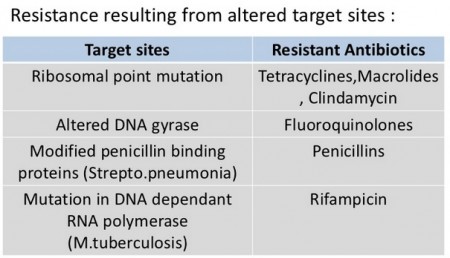
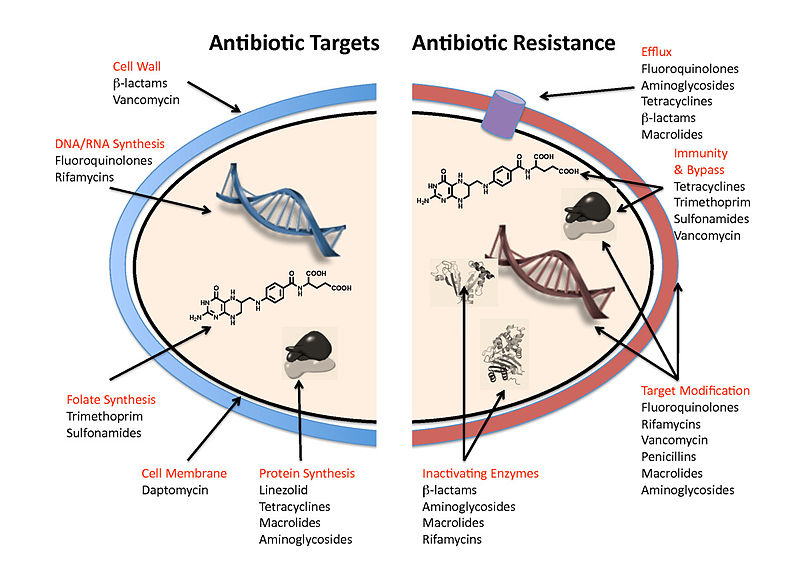
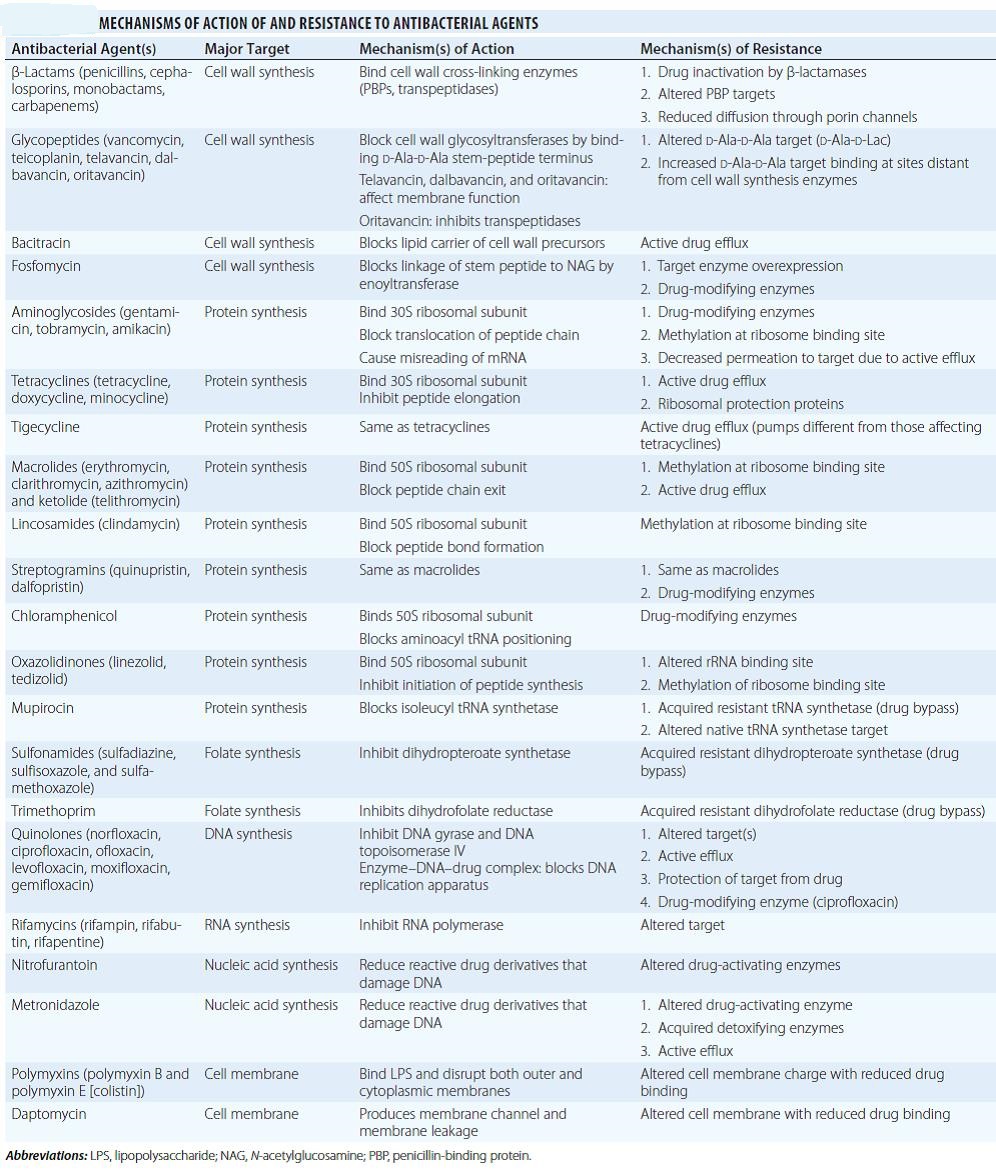
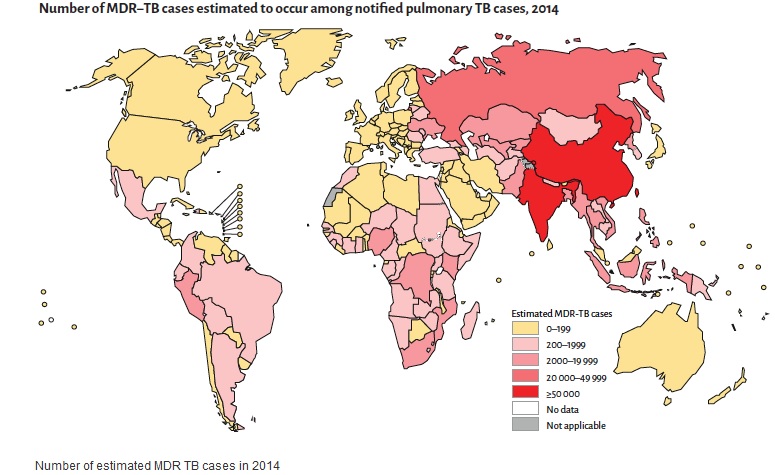
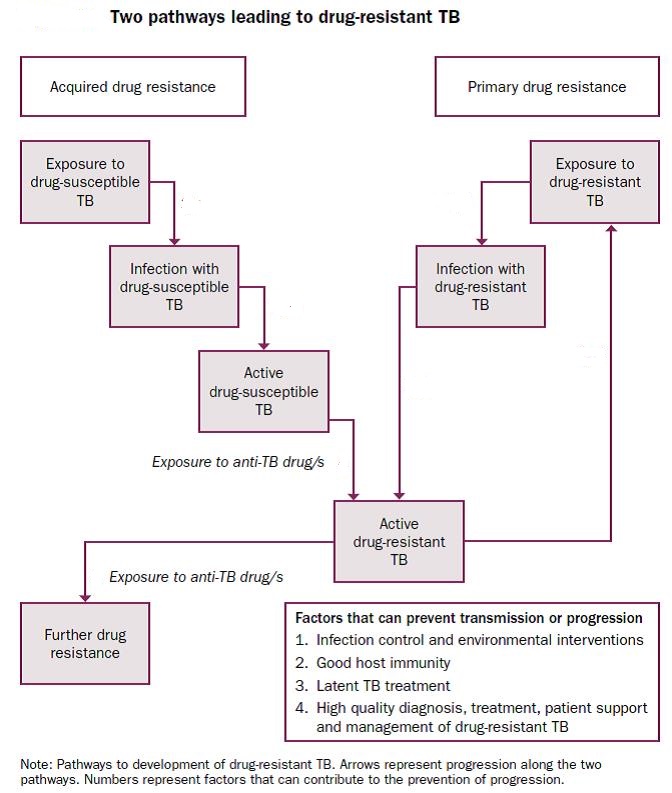
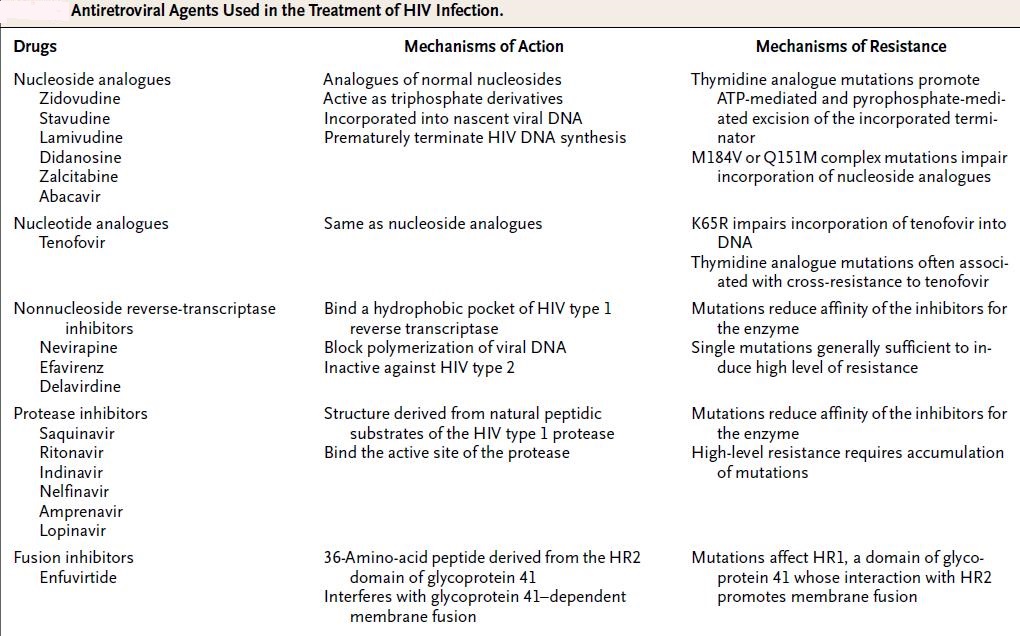
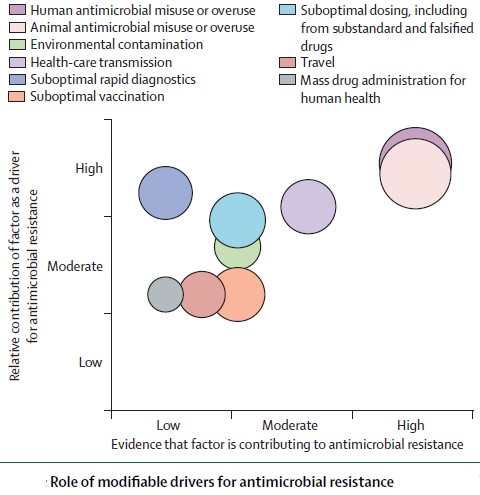

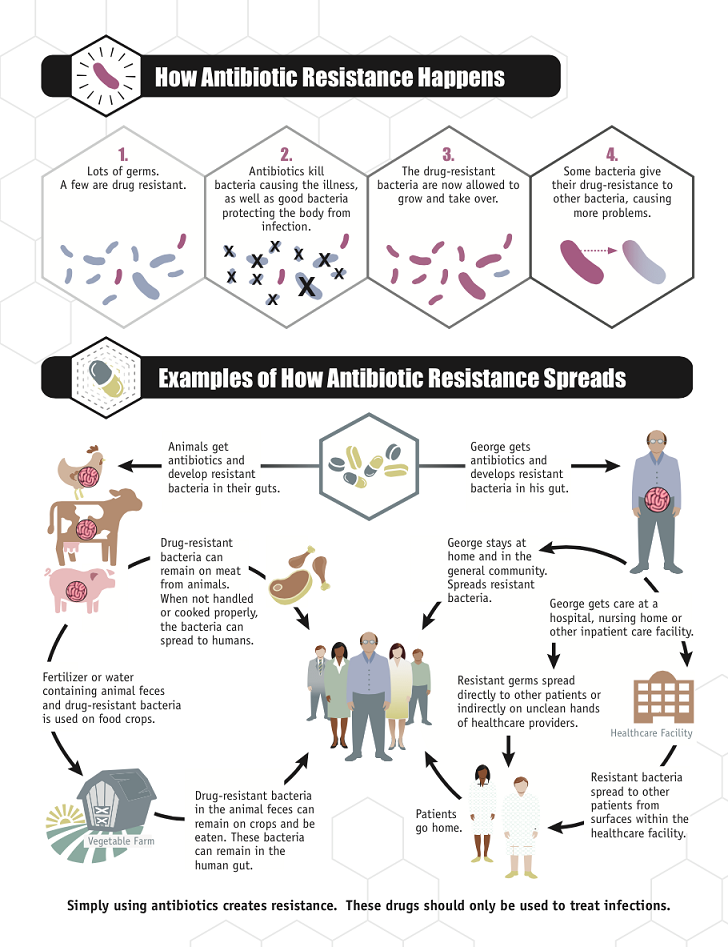
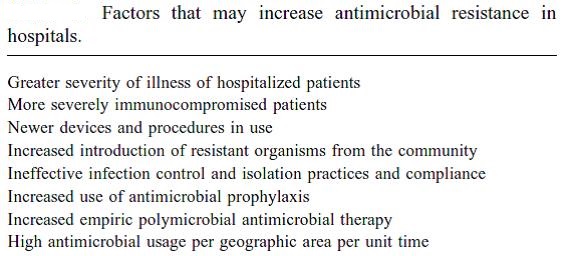
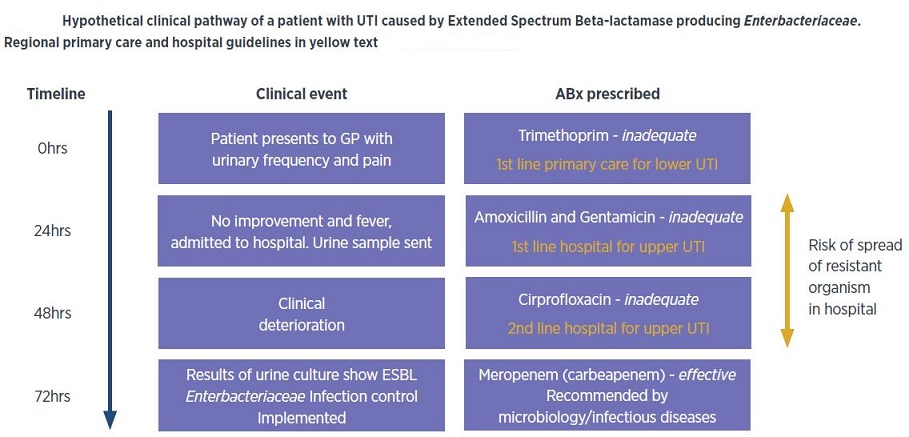
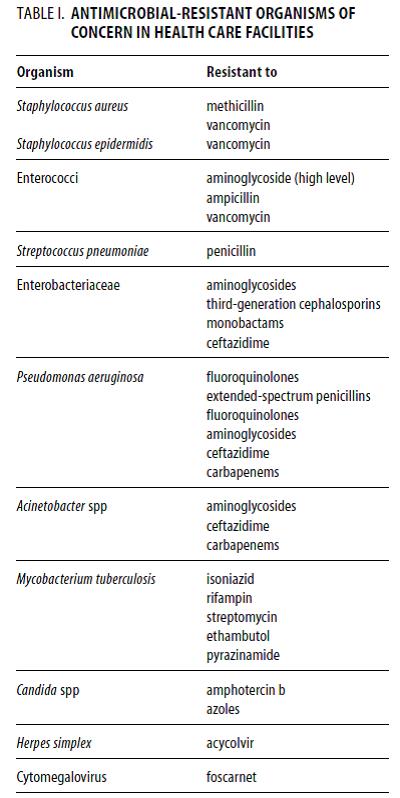
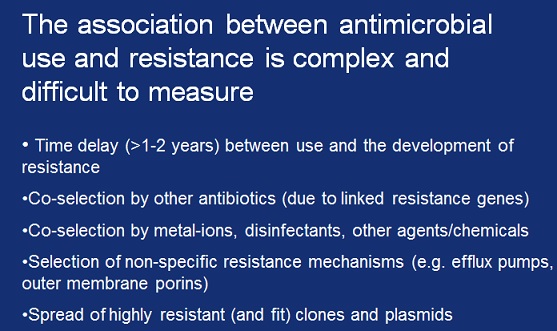
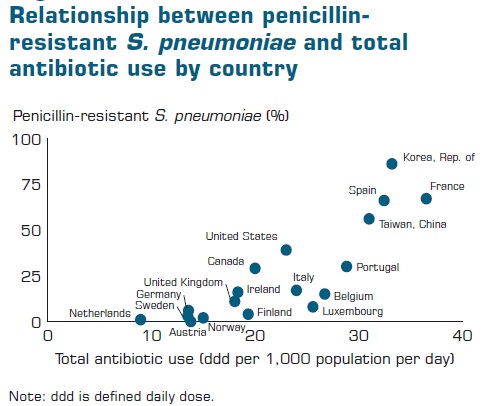
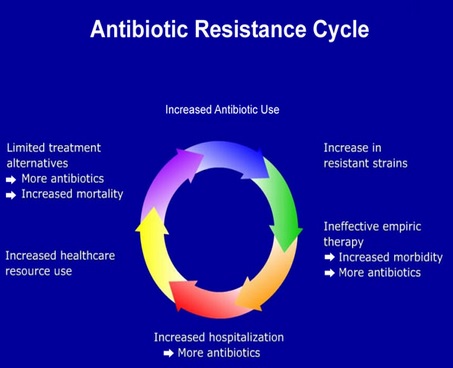
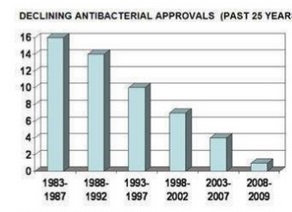
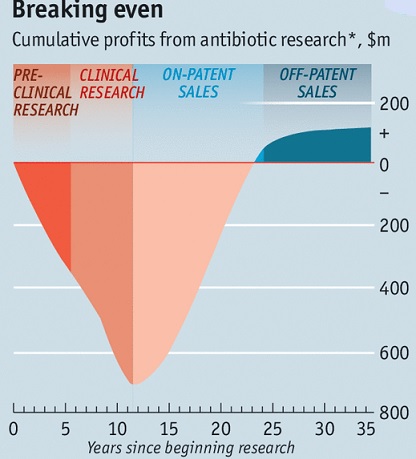
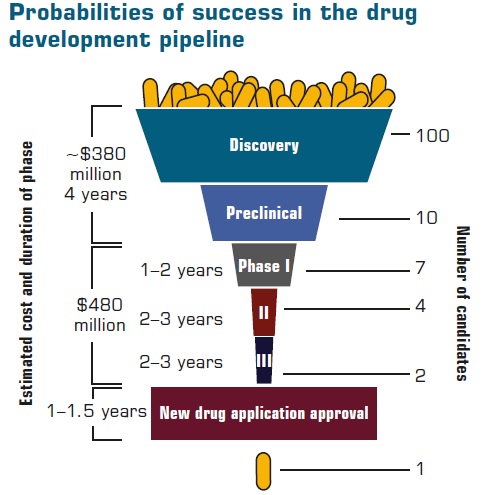
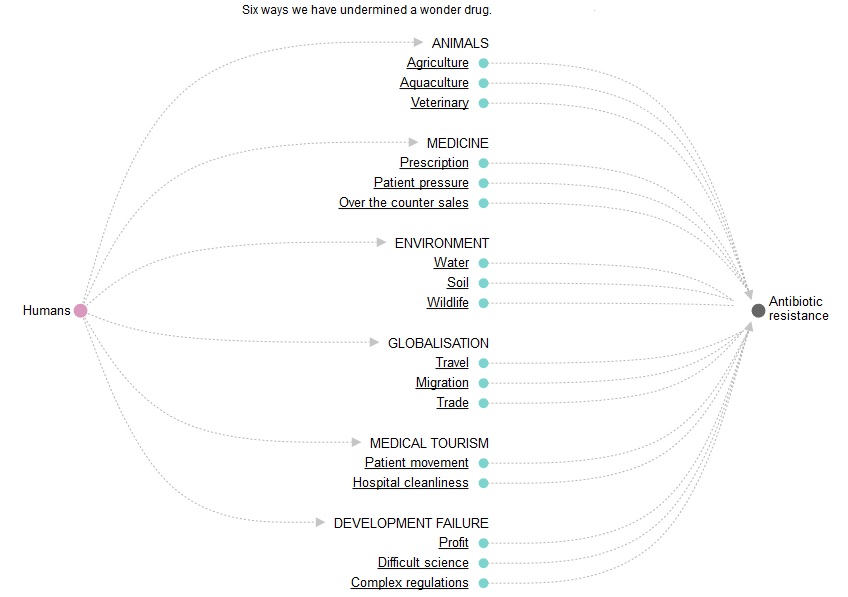
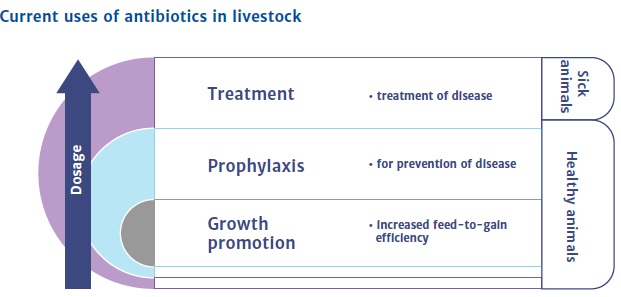
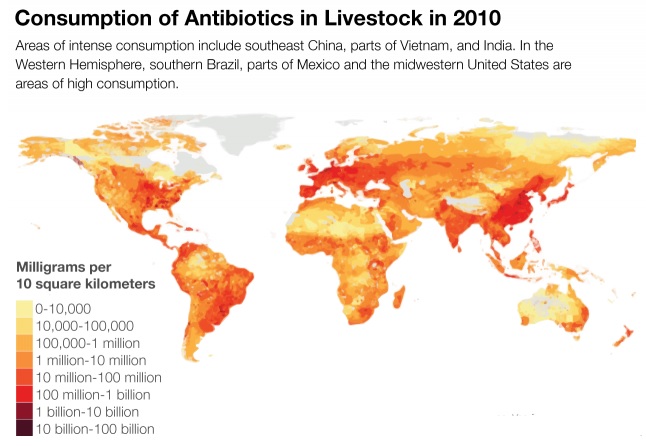
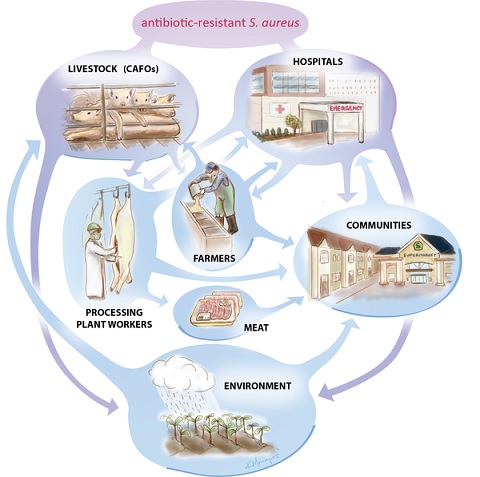
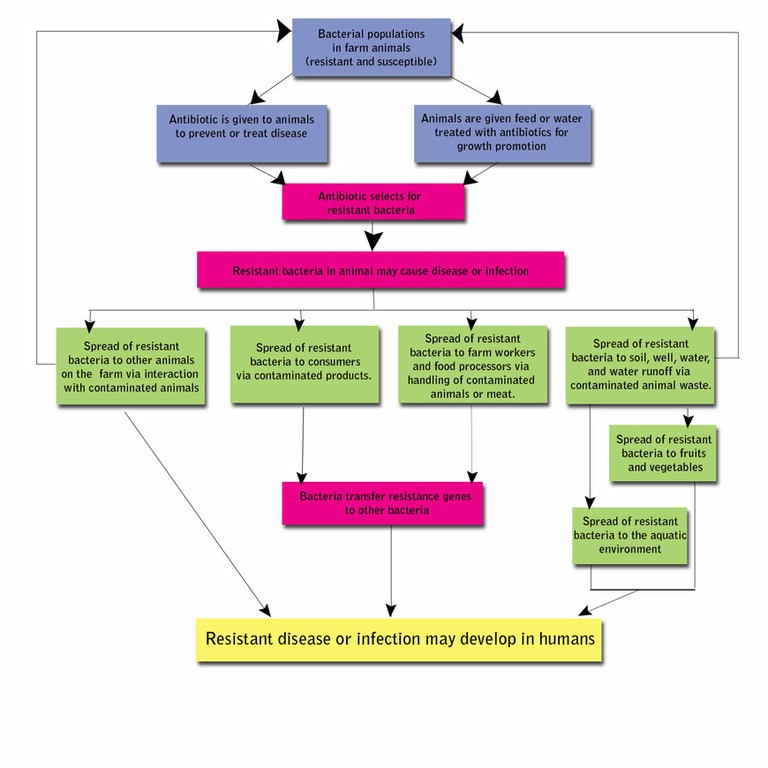
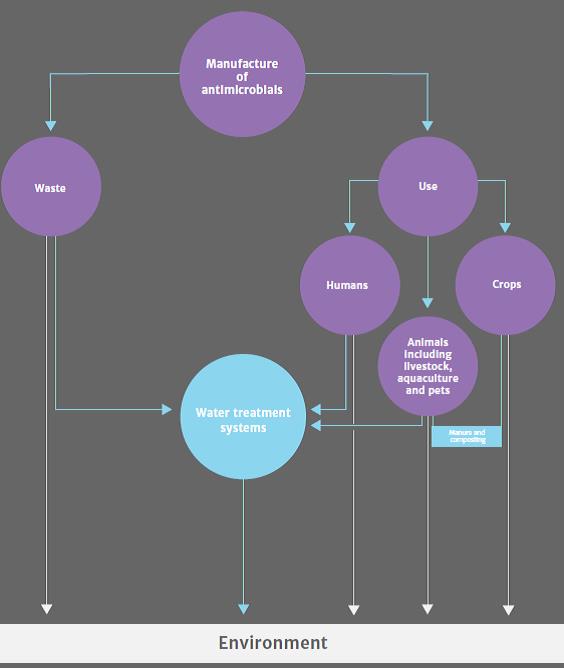
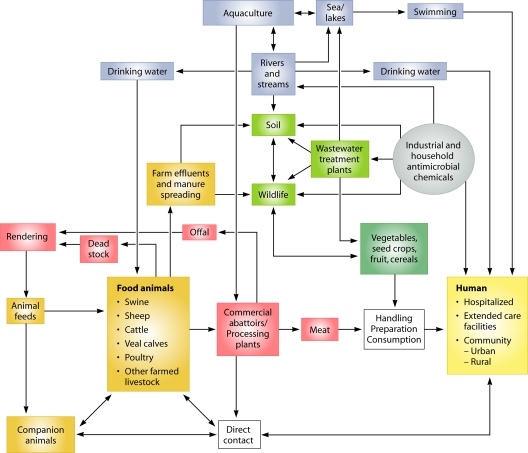
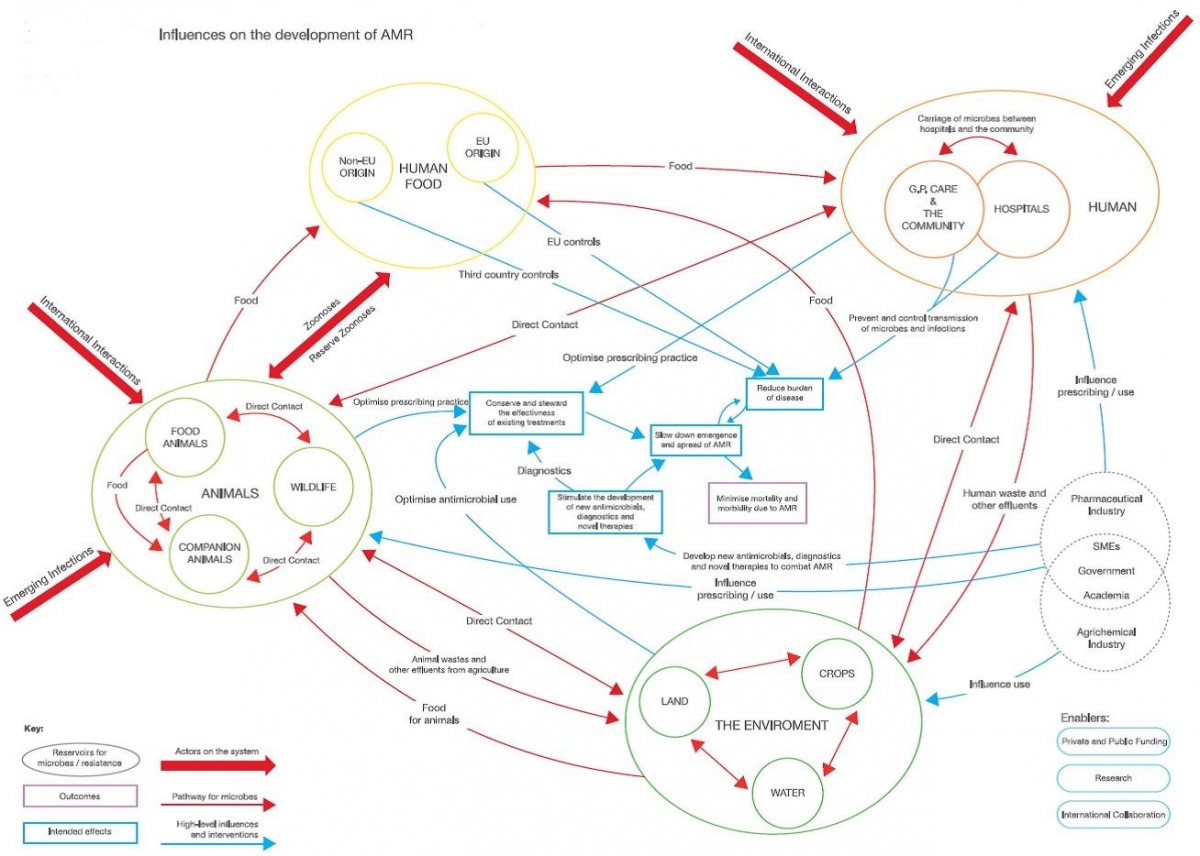
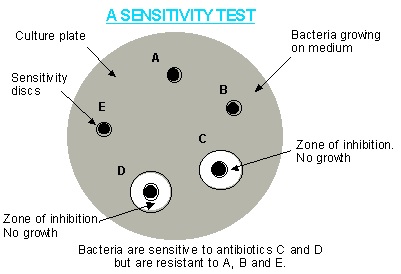
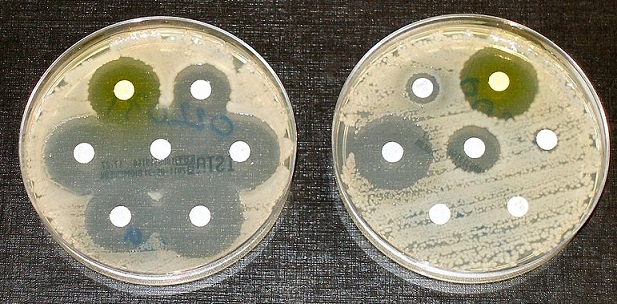
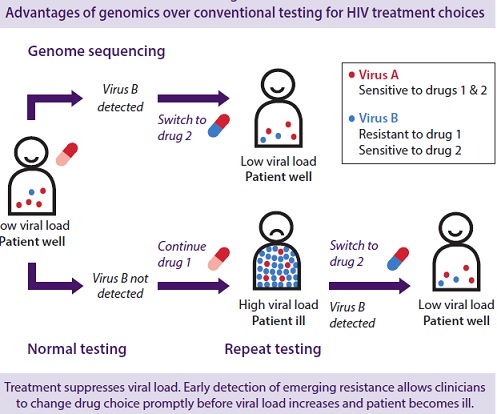
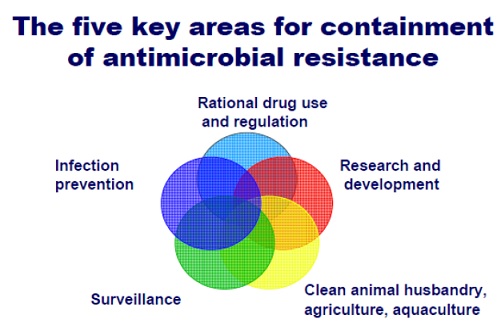

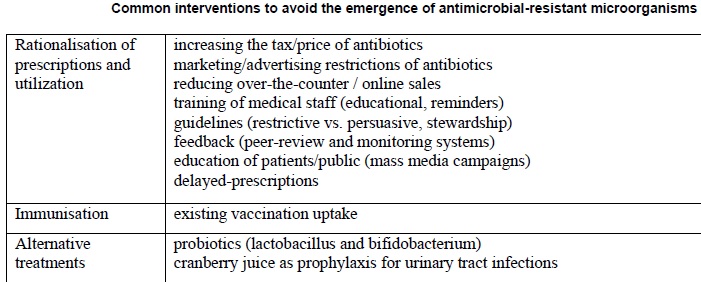
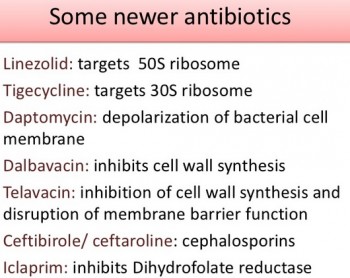
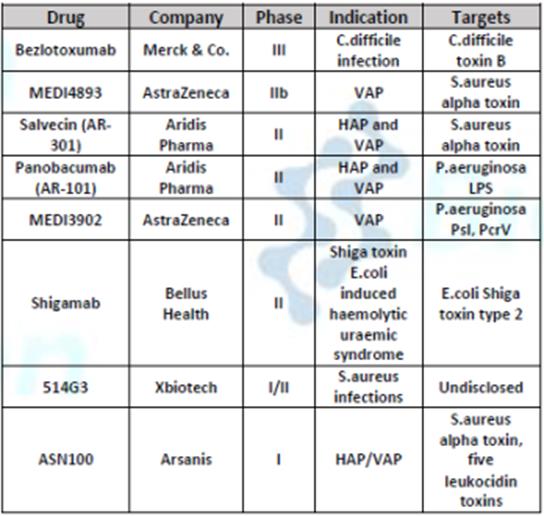
May I simpply say what a relief to find somebody who genuinely understands what they’re
discussing on the web. You actually realize how to bring a problem to light
and make it important. More and more people have to look at this and understand this side
of your story. It’s surprising you’re not more popular because you definitely
have the gift.
I couldn’t resist commenting. Exceptionally well written!
Awesome post.Really looking forward to read more. Will read on…
Great article.Thanks Again. Cool.
Undeniably believe that which you stated. Your favorite justification seemed to be on the web the easiest thing to be aware of. I say to you, I definitely get irked while people consider worries that they just don’t know about. You managed to hit the nail upon the top and also defined out the whole thing without having side effect , people could take a signal. Will likely be back to get more. Thanks|
Its such as you learn my mind! You seem to understand so much approximately this, such as you wrote the guide in it or something. I think that you can do with some percent to power the message home a bit, however other than that, that is wonderful blog. An excellent read. I will definitely be back.
Thank you ever so for you blog article.Thanks Again.
You need to be a part of a contest for one of the most useful websites on the net. I most certainly will recommend this web site!
Spot on with this write-up, I honestly think this web site needs far more attention. Iíll probably be back again to read through more, thanks for the information!
Hello my loved one! I want to say that this article is amazing, great written and include almost all vital infos. I would like to look extra posts like this .|
Terrific article
Pretty! This has been an extremely wonderful article. Many thanks for providing this info.|
excellent points altogether, you simply gained a new reader. What would you recommend about your post that you made a few days ago? Any positive?
Awesome, this is what I was looking for in bing
I consider something really special in this site.
I truly appreciate this blog.Thanks Again. Great.
Major thanks for the blog.Much thanks again. Awesome.
Thank you for your post.Really looking forward to read more. Want more.
Amazing! This blog looks exactly like my old one! It’s on a totally different topic but it has pretty much the same page layout and design. Excellent choice of colors!
I have fun with, result in I discovered just what I was having a look for. You’ve ended my 4 day long hunt! God Bless you man. Have a great day. Bye
Hi there, You’ve done a fantastic job. I will certainly digg it and personally suggest to my friends. I am sure they’ll be benefited from this website.
It is quite exciting and happy to know about such things.
Hello, Neat post. There’s an issue together with your site in web explorer, would check this… IE still is the marketplace chief and a big element of people will leave out your wonderful writing due to this problem.
Great post. I was checking constantly this blog and I’m impressed! Extremely helpful info particularly the last part 🙂 I care for such info a lot. I was looking for this certain information for a very long time. Thank you and good luck.
I love what you guys tend to be up too. Such clever
work and coverage! Keep up the very good works guys I’ve
added you guys to my blogroll.
I like this web blog so much, saved to fav. “To hold a pen is to be at war.” by Francois Marie Arouet Voltaire.
Nice read, I just passed this onto a colleague who was doing some research on that. And he actually bought me lunch since I found it for him smile Thus let me rephrase that: Thank you for lunch!
I’ve been absent for some time, but now I remember why I used to love this site. Thanks , I¡¦ll try and check back more often. How frequently you update your site?
Excellent goods from you, man. I’ve understand your stuff previous to and you are just extremely wonderful.
Enjoyed every bit of your blog.Thanks Again. Fantastic.
This is one awesome blog.Really thank you! Want more.
Thanks for the blog.Much thanks again. Want more.
Thanks for sharing, this is a fantastic blog.Thanks Again. Want more.
I truly appreciate this blog post.Really thank you! Really Great.
Fantastic blog post.Really looking forward to read more. Want more.
I really liked your article.Thanks Again. Really Great.